Abstract
Diaphanosoma excisum is the only Cladoceran in tropical freshwaters and parapatrically occurs with Diaphanosoma dubium in the transition between the tropics and subtropics. Here, we present the complete mitochondrial genome (MG) determined by next-generation sequencing and offer a phylogenetic analysis of D. excisum. The MG of D. excisum is 17,615 bp in size, including 13 protein-coding genes (PCGs), 2 ribosomal RNA, 23 tRNA, and 2 putative control regions. The MG has a biased A + T of 65.34% for base composition. Compared to D. dubium, the MG of D. excisum has one more tRNA-Met, one unknown extra putative control region and is different in the arrangement of its tRNAs. The MG sequence and tRNA order provide valuable molecular data for understanding the phylogeny and speciation of Diaphanosoma.
Diaphanosoma, the ‘tropical Daphnia’, is common and ubiquitous in the tropics and subtropics (Dumont Citation1994; Sarma et al. Citation2005; Dumont et al. Citation2021). Among Diaphanosoma species, Diaphanosoma excisum and Diaphanosoma dubium are two of the most common and dominant species. They are parapatrically distributed in warmer waters and rarely coexist in the transition zone between the tropics and subtropics (Korovchinsky et al. Citation2017; Liu et al. Citation2018; Pajk et al. Citation2018). In the tropics, D. excisum is frequently the only Cladoceran present (Kotov et al. Citation2013). In contrast to D. dubium, which is widespread in subtropical waters and with a dominance in China, D. excisum is restricted to the coastal islands of the southern part of the country (Chen et al. Citation2011). Pajk et al. (Citation2018) measured the life history traits of clones from 16 populations of D. dubium and D. excisum under a broad temperature range from 10 °C to 40 °C and showed that D. excisum had a narrower thermal performance curve (TPC) and a higher optimum temperature than the subtropical D. dubium, but failed to reproduce at ≤15 °C. Stable thermal niche difference is considered to play a critical role in shaping Diaphanosoma species range. To reveal the potential mechanism underlying niche divergence of the two congeners, there is a need to analyze not only ecological, but also genetic information. Here, we sequenced and annotated the mitochondrial genome (MG) of D. excisum and compared it to the published MG of D. dubium (Liu et al. Citation2017).
Living animals were collected from Donghu lake (110.35°E, 20.04°N) in Hainan Island, China, and mass-cultured in a small aquarium. The specimens (accession number COZOOP02002B, Ningning Liu ([email protected])) and their extracted DNA were preserved at −20 °C in the Aquatic Collection of Institute of Hydrobiology, Jinan University, Guangzhou, China. Thousands of individuals were collected and preserved in −80 °C. Genomic DNA was extracted using the TIANamp Marine Animals DNA Kit (TIANGEN BIOTECH CO., LTD (Beijing, China) and sequenced using a next-generation method on Illumina platform HiSeq 2500. The assembly and annotation procedure followed Xu et al. (Citation2018) with the COI sequence of D. excisum as the seed. The assembled sequence was annotated in MITOS WebServer (Bernt et al. Citation2013). Base depth was qualified with BamDeal (https://github.com/BGI-shenzhen/BamDeal). Transfer RNA genes were conformed with tRNAscan-SE version 2 (Lowe and Eddy Citation1997). To verify the putative control regions, we re-sequenced individual animals of D. excisum with four pairs of primers, i.e. DE1F: ggcgtgatgagatggtgaatta and DE1R: ggctgcaacaaacccataaac, DE2F: gaacggcaagacgagagaaa and DE2R: cgttgggtatcacgacagtaaa, DE3F: tcgtacgctctcgtacctatac and DE3R: gccgactttggcttcatcta, DE4F: gcgacctcgatgttggatta and DE4R: cgcatagagacacatgggtatag.
The complete MG size of D. excisum (GenBank accession number: MW476927) was 17,615 bp. It included 13 protein-coding genes (PCGs), 2 rRNAs, 23 tRNAs, and 2 putative control regions. The nucleotide composition was A + T biased with GC content of 34.66%. Most PCGs are initiated by a typical ‘ATN’ codon, while COX1 uses ‘TTG’ as the start codon. The tRNA genes have lengths ranging from 63 to 72 bp, and they are fold into a typical cloverleaf structure. The two putative control regions were 1143 and 1215 bp in length, respectively. However, one more tRNA-Met was detected in D. excisum, which was uncommon for invertebrate. The same phenomenon was also found in the MG (GenBank accession number: MT356995) of another Diaphanosoma species. The arrangement of tRNA was different for the two species, but no shift of PCGs was discovered. Besides, one more putative control region was detected in D. excisum.
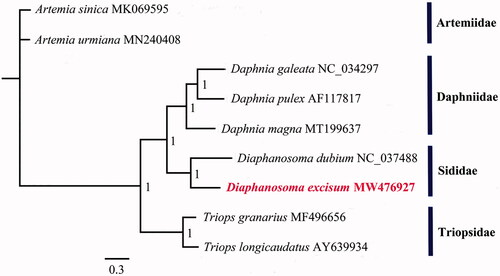
To explore the phylogenetic relationships of D. excisum and D. dubium and seven other species from three families in Brachiopoda which have complete MGs, a phylogenetic tree was obtained using Bayesian inference (BI) analysis based on entire PCGs sequences and two rRNA sequences. The Bayesian analysis was performed using MrBayes version 3.1.2 (Ronquist and Huelsenbeck Citation2003) with the GTR + G+I model of nucleotide substitution. The MGs of the two Diaphanosoma species have a similar A + T composition (65.34% for D. excisum and 65.6% for D. dubium). The phylogenetic tree shows that D. excisum was fully resolved in a clade with D. dubium in the Sididae (). The Sididae (Ctenopda) was phylogenetically closer to the anomopod Daphniidae, but divergent from Triopsidae (order Notostraca) and Artemiidae (order Anostraca).
Figure 1. Phylogenetic estimate of the position of D. excisum and eight other species of Branchiopod Crustaceans. The numbers at the nodes are posterior probabilities. The phylogeny was reconstructed based on nucleotides of 13 mitochondrial PCGs and 2 mitochondrial rRNA genes using Bayesian inference (BI) analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov) under the accession no. MW476927. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA700806, SRR13664529, and SAMN17839477, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen H, Lin QQ, Xu L, Han BP. 2011. Redescription of common species of Diaphanosoma (Cladocean, sididae) in China. Ecol Sci. 30:223–228.
- Dumont HJ. 1994. On the diversity of the Cladocera in the tropics. Hydrobiologia. 272(1–3):27–38.
- Dumont HJ, Han BP, Guo F, Chen H, Cheng D, Liu P, Xu L, Sanoamuang LO, Rietzler A, Xu S, et al. 2021. Towards a phylogeny and biogeography of Diaphanosoma (Crustacea: Cladocera). Aquat Ecol. https://doi.org/10.1007/s10452-020-09819-0
- Korovchinsky NM, Walsh EJ, Smolak R. 2017. Diaphanosoma Fischer, 1850 (Crustacea: Cladocera: Sididae) of Lake Turkana (East Africa), with the description of a new species of the genus. Zootaxa. 4250(1):77–89.
- Kotov A, Korovchinsky NM, Petrusek A. 2013. World checklist of freshwater Cladocera species. http://fada.biodiversity.be/group/show/17
- Liu P, Xu S, Huang Q, Dumont HJ, Lin Q, Han BP. 2017. The mitochondrial genome of Diaphanosoma dubium with comparison with Daphnia magna. Mitochondrial DNA Part B. 2(2):926–927.
- Liu P, Xu L, Xu S, Martínez A, Chen H, Cheng D, Dumont HJ, Han B-P, Fontaneto D. 2018. Species and hybrids in the genus Diaphanosoma Fischer, 1850 (Crustacea: Branchiopoda: Cladocera). Mol Phylogenet Evol. 118:369–378.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Pajk F, Zhang JX, Han BP, Dumont HJ. 2018. Thermal reaction norms of a subtropical and a tropical species of Diaphanosoma (Cladocera) explain their distribution. Limnol Oceanogr. 63(3):1204–1220.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Sarma SSS, Nandini S, Gulati RD. 2005. Life history strategies of cladocerans: comparisons of tropical and temperate taxa. Hydrobiologia. 542(1):315–333.
- Xu SL, Guan ZY, Huang Q, Xu L, Vierstraete A, Dumont HJ, Lin QQ. 2018. The mitochondrial genome of Atrocalopteryx melli Ris, 1912 (Zygoptera: Calopterygidae) via Ion Torrent PGM NGS sequencing. Mitochondrial DNA Part B. 3(1):115–117.
