Abstract
Bupleurum euphorbioides is a rare native plant attributed with analgesic, gallbladder-supportive, and other functions in China and the Republic of Korea. However, the complete chloroplast genome sequence of the native plant B. euphorbioides has not been determined. In this study, we sequenced the complete chloroplast genome sequence, and examined the molecular phylogeny and genetic information of B. euphorbioides. The total chloroplast genome of B. euphorbioides was 154,871 bp in length with a large single-copy region (85,089 bp), small single-copy region (17,714 bp), and pair of inverted repeats regions (26,034 bp). The chloroplast genome encoded a total of 176 genes, including 131 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic tree indicated that B. euphorbioides was the most closely related to B. latissimum.
Bupleurum spp. is one of the largest genera in family Apiaceae, with more than 150 species distributed mainly in Eurasia (Pimenov and Leonov Citation1993; Neves and Watson Citation2004). Bupleurum species are well-known for their analgesic, antipyretic, gallbladder-supportive, and other functions (Luo and Jin Citation1991). Therefore, they are very popular in traditional Chinese and Korean medicine (Luo and Jin Citation1991; Pan Citation2006; Tan et al. Citation2007). In the Republic of Korea, native Bupleurum species have been analyzed for polymorphic DNA by sequencing (Moon et al. Citation2009). Of these, B. euphorbioides has been designated as a rare plant in 1997 by the Republic of Korea Forest Service. B. euphorbioides is a native plant inhabiting the Mt. Seorak, Mt. Sobaek, and Mt. South Deogyu areas (So et al. Citation2006). However, the complete chloroplast genome of B. euphorbioides, native to the Republic of Korea, has not been sequenced.
In this study, we sequenced the complete chloroplast sequence of B. euphorbioides and determined its molecular phylogeny and genetic information. Fresh plants of B. euphorbioides were collected from Mt. Seorak, Kangwon-do, Republic of Korea (38°11′9″ N, 128°28′54″ E). A voucher specimen (TKMII-33-2) was deposited at the Medicinal Crops Seed Supply Center of the National Institute for Korean Medicine Development (NIKOM). Whole chloroplast DNA was isolated using the DNeasy Plant mini kit (Qiagen, Hilden, Germany), and the raw read sequence (9,377,516 bp) was obtained using the Illumina platform (HiSeq 2500 and NovaSeq) at Genotech Inc. (Yuseong-gu, Daejeon, Republic of Korea). Raw reads having 95% ≥ Q30 (base Phred quality score) were assembled using NOVOPlasty v2.6.7 (Dierckxsens et al. Citation2017). Raw sequencing data were registered in SRA with accession number SRX9695268. The assembled sequences were annotated using the dual organellar genome annotator (Dogma; Wyman et al. Citation2004) followed by visualization, analysis, chloroplast genome annotation, and GenBank submission using the tool CPGAVAS2 (Shi et al. Citation2019). The annotated chloroplast genome sequence was submitted to the NCBI GenBank database under the accession number MT821948. Bupleurum falcatum was used as reference genome in all processes.
The chloroplast genome of B. euphorbioides included two single-copy regions (large single-copy (LSC) and small single-copy (SSC)) and a pair of inverted repeat (IR) regions comprising 85,089 bp, 17,714 bp, and 26,034 bp, respectively. The chloroplast genome encoded a total of 176 genes, including 131 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The GC content, total LSC, total SSC, and total IR regions constituted 37.7%, 35.8%, 31.4%, and 42.9% of the chloroplast genome, respectively.
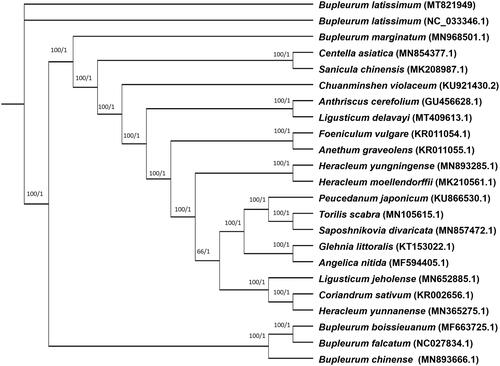
For phylogeny of B. euphorbioides, 10 complete chloroplast genome sequences belonging to Apiaceae(family) were aligned by MAFFT (Katoh and Standley Citation2013). Phylogenetic tree was designed by Maximum likelihood(ML) and Bayesian inference(BI) models and constructed via RAxML 8.2.0 (Stamatakis Citation2014) and MrBayes 3.2.7 (Huelsenbeck and Ronquist Citation2001), respectively. As a result, based on the resulting phylogenetic tree, B. euphorbioides was the most closely related to B. latissimum (). So, this analysis result could improve understanding for B. euphorbioides and provide essential data in the evolution of related groups.
Acknowledgments
We thank Dr. S. H. Cha and S.W. Lee for assisting with the nucleotide data analysis of plant materials.
Disclosure statement
The authors declare that there are no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT821948. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA685933, SRX9695268, and SAMN17101188, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Luo SQ, Jin HF. 1991. Chemical constituents of the aerial parts of six species of Bupleurum genus medicinally used in south-west region of China. Chin J Chin Mater Med. 16(6):353–356.
- Moon BC, Choo BK, Ji UN, Yoon TS, Lee AY, Cheon MS, Kim BB, Kim HK. 2009. Molecular authentication and phylogenetic relationship of Bupleurum Species by the rDNA-ITS Sequences. Korean J Herbol. 24(3):59–68.
- Neves SS, Watson MF. 2004. Phylogenetic relationships in Bupleurum (Apiaceae) based on nuclear ribosomal DNA its sequence data. Ann Bot. 93(4):379–398.
- Pan SL. 2006. Bupleurum species: scientific evaluation and clinical applications. Boca Raton: RCR Press.
- Pimenov MG, Leonov MV. 1993. The genera of the Umbelliferae: a nomenclator. Kew: Royal Botanic Gardens.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- So SK, Kim MY, Park HR, Seo EK, Kwon HJ, Song HK. 2006. Ecology of Bupleurum euphorbioides population. J Korean Soc Environ Restor Technol. 9(6):86–94.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tan LL, Hu ZH, Cai X, Cai X, Chen Y, Shi W. 2007. Histochemecal localization and the content compare of main medicinal components of vegetative organs in Bupleurum chinense DC. J Mol Cell Biol. 40(4):214–220.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.