Abstract
The complete Eclipophleps carinata mitogenome was sequenced through Illumina HiSeq 2500 platform and the resulting data were analyzed in this paper. The mitochondrial genome of E. carinata is a typical circular DNA molecule of 15,781 bp with 37 genes and 74.5% A + T content, which encoded 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and the control region. The E. carinata mitochondrial genome and 27 mitochondrial genomes (downloaded from NCBI) were employed to construct phylogenetic tree, in which Ruspolia dubia and Teleogryllus occipitails were the outgroups. Phylogenetic reconstruction validated the taxonomic status of E. carinata, which was placed in the monophyletic Gomphocerinae in Acrididae.
Eclipophleps carinata L. Mitshenko, 1968 belongs to Eclipophleps using morphometrics. It is only found in the high alpine meadows area of western Mongolia. The genetics of E. carinata have not been investigated despite the interest in studying grasshopper biodiversity of western Mongolia. Here, we present the complete mitochondrial genome of the adult E. carinata collected from Mongolia in July 2019, and the typical specimen was kept in the Inner Mongolia Normal University nature museum.
Materials and methods
The specimens in this experiment were taxonomically identified by Professor Nonnaizab from Inner Mongolia Normal University, and the E. carinata specimen was collected from Mongolia, Lake Tonkhil, Tonkhil soum, province Govi Altai (46.15131 N, 93.53942 E, 2107) in July 2019.
The total genomic DNA was extracted and sequenced using Illumina HiSeq 2500 platform, de novo assembly was conducted by SPAdes v3.10.1 (http://cab.spbu.ru/software/spades/) with K-mer auto (Boardman et al. Citation2019).
Different from the published mitochondrial genomic of the Gomphoceridae in NCBI, the location and fragment size of protein-coding genes, tRNA genes, rRNA genes and D-loop region were determined. tRNAscan-search server (http://lowelab.ucsc.edu/tRNAscan-SE/) and MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py) were applied to predict the tRNA genes secondary structure, which cannot be predicted accurately on the online website, thus it is necessary to manually predict the secondary structure of tRNA gene according to the published secondary structure of tRNA gene. The base content, sequence length and codon preference of mitochondrial genome, J-chain coding genes, N-chain coding genes were analyzed and counted in MEGA X respectively (Sudhir et al. Citation2018).
We retrieved and downloaded nucleotide sequences of the 13 protein-coding genes for 27 species of insects from the NCBI. Protein-coding genes of the E. carinata determined in the current study were added, thus generating a dataset of 28 taxa (27 Acrididae species and two outgroup species (Ruspolia dubia and Teleogryllus occipitails as outgroups)). Each protein-coding gene was aligned individually based on codon-based multiple alignments using the MAFFT (Kazutaka et al. Citation2005) algorithm within the Phylosuite (Zhang et al. Citation2020). Poorly aligned sites were removed from the protein alignment before back-translate to nucleotides. The best‐fit evolutionary model was searched by ModelFinder (Kalyaanamoorthy et al. Citation2017) algorithm within the Phylosuite. The ML phylogenetic analyses were performed by IQ-TREE v1.6.8 (Nguyen et al. Citation2015), select Ultrafast for ‘Bootstrap,’ select 5000 for ‘Num of Bootstrap,’ ensure SH-alRT test, and the default value of repeated sampling is 1000.
Results
The mitochondrial genome of E. carinata is a typical circular DNA molecule of 15,781 bp with 37 genes and biased toward A and T, with 74.4%, it included 42.9% A, 31.5% T, 15.0% C, and 10.6% G, submitted to NCBI (https://www.ncbi.nlm.nih.gov/genbank/), which was assigned the accession MN968347.
The location of the 13 PCGs is identical to that for Gomphocerippus rufus mitogenome with nine PCGs on the J chain, and four on the N chain (Sun et al. Citation2010). The A + T content of protein-coding genes, tRNA genes, rRNA genes, and control region are all higher than that of G + C, which are 73.8%, 72.2%, 76.0%, and 79.6% respectively.
Among the PCGs, ATG is the start codon among seven PCGs (ND2, COII, ATP6, COIII, ND4, ND4L, and CYTB), ATT for three (ND5, ND6, and COI), ATA for two (ND3 and ND1), and ATC for ATP8, TAA is the stop codon for 12 PCGs and TAG for ND1. The overlapping areas of ATP6/ATP8 and ND4/ND4L are both 7 bp, which is ‘ATGATAA,’ this phenomenon was also found in many invertebrates, including Brachyrhynchus hsiaoi (Li et al. Citation2016), Aradus compar, Libiocoris heissi, and Aneurus sublobatus (Song et al. Citation2016).
The mitogenome consisted of 22 tRNAs and two rRNAs. All of 22 tRNAs range from 62 bp (tRNAPro) to 71 bp(tRNAVal). The DHU arm of tRNASer(AGN) has only one base pair and can not form a typical secondary structure, there are only two bases on the DHU loop. Both rRNAs are AT rich with >70%. The 16S rRNA is 1302 bp and the 12S rRNA is 792 bp. The length of the control region is 948 bp.
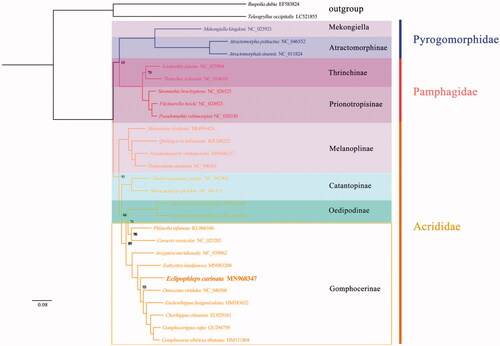
We constructed the maximum-likelihood (ML) phylogenetic tree based on the 13 PCGs of 28 species (including two outgroups from Gryllidae and Tettigoniidae). The phylogenetic tree topology structure is (Pyrgomorphidae+ (pamphagidae+ (Acrididae))) (). Eclipophleps carinata is assigned to be within the family of Gomphocerinae in Acridoidea. The phylogenetic relationship resulted from this study bearing bothsimilarities and differences in comparison to those based on morphology. Except for Gomphocerinae, all taxa in this study are monophyletic, which is supported by very high-confidence value in both trees. It is obvious that dense taxonomic sampling of Gomphocerinae is in demand to analyze the phylogenetic position of this group of grasshoppers (Zhang et al. Citation2016).
Acknowledgments
We thank Altanchimeg Dorjsuren who works in Institute of Biology, Mongolian Academy of Sciences and provided the Eclipophleps carinata specimen.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MN968347. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA685974, SRR13269645, and SAMN17102428 respectively.
Additional information
Funding
References
- Boardman L, Eimanifar A, Kimball R, Braun E, Fuchs S, Grünewald B, Ellis JD. 2019. The mitochondrial genome of Apis mellifera simensis (Hymenoptera: Apidae), an Ethiopian honey bee. Mitochondrial DNA B Resour. 5(1):9–10.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kazutaka K. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Li H, Shi A, Song F, Cai W. 2016. Complete mitochondrial genome of the flat bug Brachyrhynchus hsiaoi (Hemiptera: Aradidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):14–15.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Song F, Li H, Shao R, Shi A, Bai X, Zheng X, Heiss E, Cai W. 2016. Rearrangement of mitochondrial tRNA genes in flat bugs (Hemiptera: Aradidae). Entific Rep. 6:25725.
- Sudhir K, Glen S, Michael L, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Sun H, Zheng Z, Huang Y. 2010. Sequence and phylogenetic analysis of complete mitochondrial DNA genomes of two grasshopper species Gomphocerus rufus (Linnaeus, 1758) and Primnoa arctica (Zhang and Jin, 1985) (Orthoptera: Acridoidea). Mitochondr DNA. 21(3–4):115–131. 10.3109/19401736.2010.482585
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. Phylosuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhang Y, Liu B, Zhang H, Yin H, Zhang D. 2016. The complete mitochondrial genome of Pacris xizangensis (Orthoptera: Acridoidea: Gomphoceridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):320–321.