Abstract
Trichoptera are a group of the benthic organism, almost all of which live in water during their life cycle. Trichoptera usually develop through egg, larva, pupa, and moth stages. In its larval stage, Trichoptera usually live in water and are often called the caddisfly. In this study, the mitochondrial genome of Macrostemum floridum was analyzed. The total length of the mitochondrial genome is 15,424 bp and consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and one control region. The genome has a typical mitochondrial gene sequence of Trichoptera. Phylogenetic analysis of the mitochondrial genomes of 23 species of Trichoptera and Lepidoptera showed that M. floridum forms a monophyletic group with other species of Lepidoptera.
Macrostemum floridum larva is a medium-sized caddisfly. The whole body is green and the head and chest are bony and oval in shape. The thorax is well developed and there are no gastropods in its abdomen, but there are many brushes, which are used to sense the direction and velocity of the external water flow. M.acrostemum floridum larvae spin silk and form funnel-shaped nets in water to filter food in water, including algae, fungi, and organic debris (Wallace et al. Citation1977; Long et al. Citation2000). They prefer to live in clean rivers without pollution and M. floridum also have certain requirements for water velocity and depth. Therefore, M. floridum can be used as an indicator organism to detect and evaluate the water quality (Wallace and Webster Citation1996; Li and Zhou Citation2001; Long and Zhang Citation2002; Huang and Cai Citation2006; Chen Citation2013; Lin et al. Citation2017).
Mitochondria are important markers widely used in the study of genetic diversity, species origin and evolution, and molecular taxonomy. By comparing groups associated with the system development between the change of the length of the mitochondrial genome, tRNA anticodon, or the change of secondary structure, start and stop codon, base composition, preferences, using type, and frequency of codon and rearrangement of the mitochondrial genome characteristics can be for the evolution of different species were analyzed, and determine the evolution of gene sequence (Pan and Bu Citation2005).
In this study, the complete mitochondrial genome M. floridum was sequenced and analyzed, which is helpful to identify the species and locate the evolutionary relationship of M. floridum, so that it can be protected and used from a biological perspective.
M.acrostemum floridum samples used in this study were collected from Shiwanda Mountain in Guangxi Province, China (108.26E, 22.11N). A specimen has been deposited in the Ocean College Marine Specimen Showroom of Beibu Gulf University of China (voucher No. BBGC 00013). The mitochondrial genome of M. floridum was sequenced and assembled using Illumina high-throughput sequencing technology and SPAdes 3.5.0 version software (http://cab.spbu.ru/software/spades/). MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) and ORF (https://www.ncbi.nlm.nih.gov/orffinder/) Finder were used to annotate the mitochondrial genome. The preliminary results were compared with the protein-coding and ribosomal RNA of the mitochondrial genome of related species using BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and BLASTn methods.
The total length of the mitochondrial genome is 15,424 bp, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and one control region. The base composition of the H chain was 39.35% A, 39.52% T, 6.35% G, and 14.78% C, which is similar to that of other Trichoptera insects, and the AT content was higher than that of GC. All tRNA genes except tRNA-Ser (TCT) could be folded into a typical cloverleaf structure with a length of 60–73 bp. 16 s RNA and 12 s RNA genes were located between tRNA-Val and tRNA-Leu and between tRNA-Val and the D-loop, respectively. The initiation codons of 13 protein-coding genes were ATN. The stop codon of the protein-coding genes was an incomplete termination codon T in cox1, cox2, and nad6, and ATN in the remaining genes.
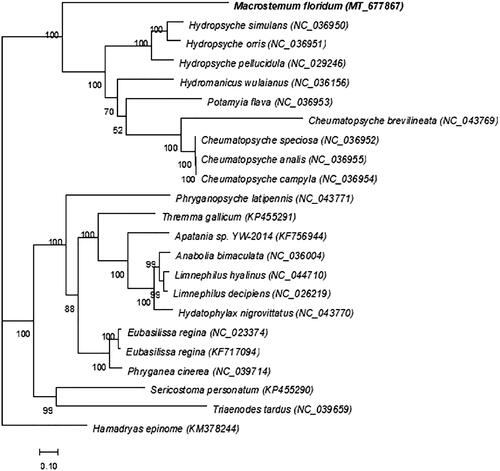
We downloaded 20 genomes of related species from NCBI, including complete or nearly complete mitochondrial genomes (nearly complete refers to complete coding genes) and newly sequenced genomes using RAxML version 8.1.5 software (https://sco.h-its.org/exelixis/web/software/raxml/index.html). The maximum likelihood (ML) method was used to construct a phylogenetic tree and the bootstrap value was set to 1000. Macrostemum floridum, along with Hydropsyche, Cheumatopsyche, Potamyia, and Hydromanicus are all a genus of the Hydropsychidae family. According to the phylogenetic tree of M. floridum, it can be confirmed that M. floridum, Hydropsyche, Cheumatopsyche, etc., are all on the same large branch, but M. floridum is different from them, forming a single branch. There is a monophyletic group in the family that includes M. floridum, and Phylogenetic analysis showed that M. floridum formed sister groups with Hydropsyche and Cheumatopsyche ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT677867.
Additional information
Funding
References
- Chen J. 2013. The three giants of water quality index biology of mayfly, stonefly and stone moth. For Hum. 33(10):82–87.
- Huang XQ, Cai DC. 2006. Application of aquatic insects in biological monitoring and evaluation of water quality. J South Chin Trop Agric Univ. 2006(02):72–75.
- Li ZP, Zhou YM. 2001. Trichoptera larvae, an important indicator of water quality monitoring. Heilongjiang Environ Bull. 14(01):77–78.
- Lin QF, Chen HF, Billy KY, Zou Y, Li XZ, Wang YJ. 2017. Acute Toxicity of Cu2 + and Pb2 + on Steaopsyche marmorata Larvae. Anhui Agric Sci. 45(18):51–53.
- Long JG, Zhang JY, Peng NN. 2000. Biological characteristics of Lepidoptera longicornis. Acta Hydrobiol. 24(4):399–401.
- Long JG, Zhang JY. 2002. Ecological characteristics of Lepidoptera longicornis. J Ecol. 21(3):25–28.
- Pan B, Bu W. 2005. Advances in the genetics and evolution of mitochondrial genomes. Chin J Biol. 2005(08):5–7.
- Wallace JB, Webster JR. 1996. The role of macroinvertebrates in stream ecosystem function. Annu Rev Entomol. 41(1):115–l39.
- Wallace J, Webster J, Woodall W. 1977. The role of filter feeders in flowing waters.Arch Hydrobiol. 79(4):506–532.