Abstract
The complete chloroplast genome of Clematis henryi var.ternata was determined in this study. The genome was 159,675 base pair (bp) in length, containing a large single-copy (LSC) region of 79,443 bp, a small single-copy region (SSC) of 18,100 bp and a pair of inverted repeats (IRs) of 31,066 bp. It contains 130 unique genes, including 86 protein-coding genes, 36 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The GC content of the complete chloroplast genome sequence was 38.0%. Phylogenetic analyses using complete chloroplast genomes showed that Clematis henryi var.ternata is most closely related to Clematis guniuensis (NC_050373.1).
There were about 300 species in the genus Clematis L. It is distributed on all continents, mainly in tropical and subtropical regions, as well as in cold regions (Tamura Citation1995; Johnson Citation1997; Wang and Li Citation2005). There were about 108 species in China, which were distributed throughout the country, especially in the southwest (Wang and Li Citation2005). Among them, Clematis henryi var.ternata is produced in the southwestern of Shannxi province, China. In this study, we analyzed the characters of the complete chloroplast genome sequence for Clematis henryi var.ternate to confirm its phylogenetic position and evolutionary relationship between the Clematis henryi var.ternata and other Ranunculaceae species.
Clematis henryi has the functions of promoting qi and relieving pain, promoting blood circulation and reducing swelling. As a traditional herbal medicine, Clematis henryi has been widely used in China for hundreds of years for the treatment of infectious and inflammatory disorders. Pharmacological studies have shown that Clematis henryi has obvious analgesic and sedative effects, and has analgesic effects on head, stomach, abdominal muscles and joint pain (Liu Citation2007). In this article, we de novo assembled the complete chloroplast genome of Clematis henryi var.ternata, which will provide genomic data for the related studies and a phylogenetic tree was generated to reveal its relationship with other Clematis species.
The samples of Clematis henryi var.ternata were collected from Ningshan County, Shannxi Province, China (33°32′28.01″N, 108°37′39.59″E). The voucher specimen was preserved at XBGH (The Herbarium of Xi’an Botanical Garden) (Voucher number: Xun Lulu et al.00361). Total genomic DNA was extracted using the modified CTAB method (Doyle and Doyle Citation1987) and TruSeq DNA Sample Prep Kit (Illumina, USA) was used to construct a genomic library consisting of an insert size of 350 bp. Sequencing was carried out on an Illumina NovaSeq 6000 platform. Raw PE reads of about 2.23 Gb were trimmed and high quality PE reads of about 2.21 Gb were subjected to de novo assembly. The NOVOplasty (Dierckxsens et al. Citation2017) were used for the de novo assembly of chloroplast genome. The sequence was annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html), and visually checked in Geneious v8.0.2 (Kearse et al. Citation2012) using the chloroplast genome of Clematis guniuensis (GenBank accession number (QMS50675.1)) as reference. Finally, a complete chloroplast genome of Clematis henryi var.ternata was obtained and submitted to Genbank (GenBank accession number (MW380948)).
The complete chloroplast genome of Clematis henryi var.ternata (GenBank accession number: (MW380948)) was 159,675 bp in length with 38.0% GC contents, consisting of a pair of inverted repeat regions (IRa and IRb) with same length (31,066 bp) separated by the large single copy (LSC, 79,443 bp) and small single copy (SSC, 18,100 bp) regions. A total of 130 genes were identified, including 86 protein-coding genes, eight ribosomal RNA (rRNA) genes, and 36 transfer RNA (tRNA)genes.
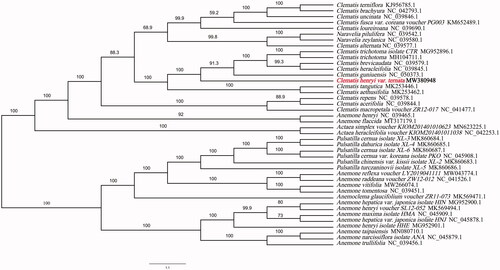
Phylogenetic tree was constructed using IQTREE v1.6.7(Nguyen et al. Citation2015) for Clematis henryi var.ternata and 41 Ranunculaceae species chloroplast genomes, with the best selected TVM + F+R4 model and 5000 bootstrap replicates. The chloroplast genome of the Clematis henryi var.ternata showed the closest relationship to that of previously reported Clematis guniuensis (NCBI reference sequence ID NC_050373.1, 159,682 bp in length) (Jiang et al. Citation2020) (). The newly characterized Clematis henryi var. ternata complete chloroplast genome would provide essential data for further study on the phylogeny and evolution of Clematis L. and provide useful resources for better understanding the physiology and evolution of Ranunculaceae. The phylogenetic tree illuminates the phylogenetic relationships among Clematis species.
Figure 1. Phylogenetic tree showing the relationship between Clematis henryi var.ternata and 41 Ranunculaceae species. Phylogenetic tree was constructed based on the complete chloroplast genomes using maximum likelihood (ML) with 5000 bootstrap replicates. Numbers in each the node indicated the bootstrap support values.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov) GenBank with the accession number (MW380948).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de Novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jiang M, Wang J, Zhang H. 2020. The complete plastome sequence of Clematis guniuensis (Ranunculaceae), a new plant species endemic to China. Mitochondrial DNA B Resour. 5(1):408–409.
- Johnson M. 1997. Släktet Klematis. Södertälje: M. Johnsons plantskola AB; p. 2–23.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data . Bioinformatics. 28(12):1647–1649.
- Liu S. 2007. Study on chemical composition of Clematis henryi Olive. Chin Tradit Patent Med. 9:1379–1380.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tamura M. 1995. Clematis L. In: Heipko P, editor. Engler’s Die Natürlichen Pflanzenfamilien. 2nd ed. Berlin: Duncker & Humblot; p. 368–387.
- Wang WT, Li LQ. 2005. A new system of classification of the genus Clematis (Ranunculaceae). Journal of Systematics and Evolution. 43(5):431–488.
