Abstract
Polyopes lancifolius is a species of Halymeniales, the fifth species-rich order within Rhodophyta. Using next-generation sequencing techniques, we recovered the complete mitochondrial genome of P. lancifolius, i.e. total 26,142 bp in length with 31% GC contents. A total of 49 functional genes were annotated, including 24 protein-coding, 23 transfer RNA, and 2 ribosomal RNA genes. The gene content and synteny have been highly congruent to those of the other halymenialean species, such as Grateloupia taiwanensis, G. filicina, and Grateloupia angusta. Interestingly, the cox1 intron and intronic Open Reading Frame (ORF) are absent in P. lancifolius, that are existed in the other three halymenialean species.
Polyopes lancifolius (Harvey) Kawaguchi & Wang is one of the introduced species in Europe originated from the northwest Pacific region (Mineur et al. Citation2010). This species is a member of Halymeniales, which is the fifth species-rich order (359 spp) within Rhodophyta (Guiry and Guiry Citation2020). However, only three mitochondrial genomes have been published in the order Halymeniales, i.e. Grateloupia angusta (KC875853; Kim et al. Citation2014), Grateloupia taiwanensis (KM999231; DePriest et al. Citation2014), and Grateloupia filicina (MG598532; Li et al. Citation2018). In this study, we sequenced and analyzed the complete mitochondrial genome of P. lancifolius, which is the first mitochondrial genome of the genus.
The specimens were collected on 25 January 2013 from Ganggu, Korea (36°21′29.6″N, 129°23′30.8″ E) and identified by rbcL phylogeny from the other halymenialean species. The voucher specimen (SKKU51) was deposited in Sungkyunkwan University (contact person: Su Yeon Kim, [email protected]). Total genomic DNA was extracted using LaboPass™ Tissue Genomic DNA Isolation Kit Mini (Hokkaido System Science Co., Ltd., Sapporo, Japan) following manufacturer’s instruction. Ion Torrent PGM (Thermo Fisher Scientific, Waltham, MA) was applied for DNA sequencing using Ion PGM Template OT2 200 Kit and PGM Sequencing OT2 200 Kit. Total reads from genome data were assembled using CLC de novo assembler implemented in CLC Genomics Workbench version 6.5.1 (Aarhus, Denmark) (https://digitalinsights.qiagen.com). Candidate mitochondrial sequences were sorted from assembled contigs by comparing with reference halymenialean mitochondrial genes. The CDS and RNA genes were manually annotated by BLAST search using the NCBI nucleotide and protein database (nt, nr) and tRNA genes were predicted by tRNA-scanSE (Lowe and Chan Citation2016) and ARAGORN (Laslett and Canback Citation2004). After the annotation, the complete genome of P. lancifolius was submitted to GenBank (accession number MW292567).
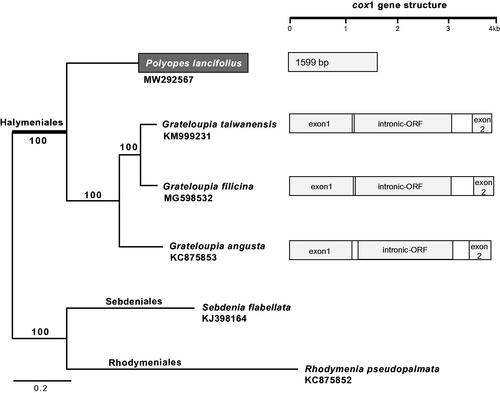
The complete mitochondrial genome of P. lancifolius is 26,142 bp in length with 31% GC contents. The overall nucleotide composition is: 9434 bp of A (36.1%), 8597 bp of T (32.9%), 3968 bp of C (15.2%), and 4133 bp of G (15.8%). The genome is comprised 49 genes, including 24 protein-coding, 23 transfer RNA, and 2 ribosomal RNA genes. The newly constructed mitochondrial genome has a highly conserved gene synteny with three other halymenialean species, such as G. taiwanensis, G. filicina, and G. angusta. However, genomic differences were identified in the absence of cox1 intron and the number of tRNA genes. All species within the order Halymeniales have a trnI (tRNA Ile) intron, whereas cox1 intron and intronic Open Reading Frame (ORF) were found only in the three Grateloupia species, except for P. lancifolius. The total number of tRNAs of P. lancifolius is 23, which is higher than that of G. angusta (19) and lower than G. taiwanensis (24) and G. filicina (24).
Total six mitochondrial genomes were selected to infer the phylogenetic relationships of P. lancefolius within the order Halymeniales. Rhodymenia pseudopalmata (KC875852) and Sebdenia flabellata (KJ398164), which are known as the sister orders of Halymeniales (Lee and Kim Citation2019; Yang et al. Citation2016), were used as outgroups. The maximum likelihood (ML) method was used to infer the phylogenetic relationship, and the tree was constructed using RAxML program (Stamatakis Citation2006). The best ML tree based on 24 CDS combined data (total 17,572 bp) fully supported (100% ML bootstrap support) the monophyly of the order Halymeniales and interspecies relationships within the order (). Considering the current taxonomical classification and our mitochondrial genome phylogeny, only three Grateloupia species have a cox1 intron and intronic ORF within the order Halymeniales (). The complete mitochondrial genome would contribute to understandings on the cox1 intron evolution in Rhodophyta.
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and writing of the article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW292567. The associated SRA and Bio-Sample numbers are PRJNA690127 and SAMN17244873, respectively.
Additional information
Funding
References
- DePriest MS, Bhattacharya D, López-Bautista JM. 2014. The mitochondrial genome of Grateloupia taiwanensis (Halymeniaceae, Rhodophyta) and comparative mitochondrial genomics of red algae. Biol Bull. 277:191–200.
- Guiry MD, Guiry GM. 2020. AlgaeBase. World-wide electronic publication. Galway, Ireland: National University of Ireland2020 Nov 3. http://www.algaebase.org. searched
- Kim SY, Yang EC, Boo SM, Yoon HS. 2014. Complete mitochondrial genome of the marine red alga Grateloupia angusta (Halymeniales). Mitochondrial DNA. 25(4):269–270.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Lee HW, Kim MS. 2019. Female reproductive structures define the novel genus, Nesoia (Halymeniaceae, Rhodophyta). Eur J Phycol. 54(1):66–77.
- Li Y, Meinita MDN, Liu T, Chi S, Yin H. 2018. Complete sequences of the mitochondrial DNA of the Grateloupia filicina (Rhodophyta). Mitochondrial DNA B Resour. 3(1):76–77.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Mineur F, De Clerck O, Le Roux A, Maggs CA, Verlaque M. 2010. Polyopes lancifolius (Halymeniales, Rhodophyta), a new component of the Japanese marine flora introduced to Europe. Phycologia. 49(1):86–96.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Yang EC, Boo SM, Bhattacharya D, Saunders GW, Knoll AH, Fredericq S, Graf L, Yoon HS. 2016. Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci Rep. 6:21361.