Abstract
In this study, the complete mitochondrial genome of Asclepios apicalis was sequenced and assembled, which was first reported in Asclepios. The mitogenome of Asclepios apicalis was 15,391 bp in length, and it contained 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, 13 protein-coding genes (PCGs), and a control region (D-loop), the overall base nucleotide compositions encoded was 42.9% A, 14.3% C, 10.0% G, and 32.8% T.
Keywords:
Asclepios apicalis (Esaki 1924) is a water bug that belongs to the family Gerridae. The water bugs conquered water surfaces worldwide and diversified to occupy ponds, streams, lakes, and even oceans. Their hairs allow them to maintain their body weight at the surface of the water and protect them from moisture and drowning (Finet et al. Citation2018). In addition, the legs of these insects are equipped with various grooming combs that contribute to cleaning and tidying the hair layers so as to obtain optimal functional efficiency. Water bugs usually play an important role in freshwater ecosystems, and information about them is essential for the study of water biology and the proper management of aquatic habitats (Cordeiro and Moreira Citation2015; Havemann et al. Citation2018). However, there is still no study on the complete mitogenome sequences for A. apicalis. Hence, it is crucial to determine the whole mitogenome of A. apicalis for further study.
The specimen of A. apicalis was collected from Kunming city, Yunnan Province (102.73°E, 25.04°N), China. The voucher specimen was deposited in Institute of Entomology, College of Life Sciences, Nankai University (Ying Cui, [email protected]), under the voucher number of A20060702. The collected specimen was preserved in 95% ethanol at −20 °C until DNA extraction. Total genomic DNA was extracted from muscle tissue of the specimen using the CTAB method (Reineke et al. Citation1998). Polymerase chain reactions (PCRs) were performed with TaKaRa LA PCR Kit Ver. 2.1 following the manufacturer's recommendations. The PCR products of the Gel electrophoresis were purified in 0.7% agarose gel and then both strands were sequenced with primer walking by Beijing Sunbiotech Co. Ltd. (Beijing, China). BioEdit 7.0 was used for sequence alignment and assembly. The characters of base composition and distribution were analyzed using Geneious Prime 2020 software (Kearse et al. Citation2012). The mitogenome was annotated using MITOS web server (Yu et al. Citation2019), and was manually checked and adjusted the annotation using Aquarius paludum (GenBank accession number: NC012841) as the reference sequence. The online tRNAscan-SE search server (Lowe and Chan Citation2016) was used to annotate the transfer RNA (tRNA) gene to determine its position, and the parameter settings were default. The start and stop codons of protein-coding genes (PCGs) were manually adjusted to fit open reading frames. The maximum-likelihood (ML) methods were used to construct the phylogenetic tree. The ML trees were obtained with 1000 bootstrap replications using MEGA X (Kumar et al. Citation2018).
The results revealed that the total length of mitogenome of A. apicalis was 15,391 bp, with the base composition of 42.9% for A, 14.3% for C, 10.0% for G, 32.8% for T, and with a high A + T content of 75.7%. The length of the non-coding region was 803 bp, which accounts for 5.2% of the total length. The length of the coding region was 14,588 bp and it contained 37 coding genes (22 tRNA genes, two ribosomal RNA (rRNA) genes, and 13 PCGs).
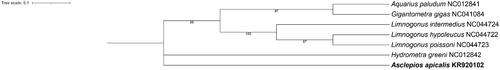
To assess the phylogenetic position of A. apicalis, the ML phylogenetic tree was constructed using the complete mitogenome sequence of Gerridae. The complete mitogenome sequence of Hydrometra greeni was set as the outgroup (Hua et al. Citation2009). The results showed that A. apicalis formed an independent lineage in the family Gerridae (). Overall, the complete mitogenome of A. apicalis can contribute to further phylogenetic study within Gerridae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study is openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number KR920102. The raw data of sequencing are openly available in Figshare at https://figshare.com/s/b7a0b835184c0e161da3.
Acknowledgements
We thank Dr Wenjun Bu (Nankai University) for the help of providing materials and sequencing. This project was supported by the Science & Technology Development Fund of Tianjin Education Commission for Higher Education (Project No. 2018KJ009).
References
- Cordeiro I, Moreira F. 2015. New distributional data on aquatic and semiaquatic bugs (Hemiptera: Heteroptera: Gerromorpha & Nepomorpha) from South America. Biodivers Data J. 3:e4913.
- Finet C, Decaras A, Armisén D, Khila A. 2018. The Achaete–Scute complex contains a single gene that controls bristle development in the semi-aquatic bugs. Proc R Soc B. 285(1892):20182387.
- Havemann N, Gossner MM, Hendrich L, Morinière J, Niedringhaus R, Schäfer P, Raupach MJ. 2018. From water striders to water bugs: the molecular diversity of aquatic Heteroptera (Gerromorpha, Nepomorpha) of Germany based on DNA barcodes. PeerJ. 6:e4577.
- Hua J, Li M, Dong P, Cui Y, Xie Q, Bu W. 2009. Phylogenetic analysis of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha): evidence from mitochondrial genomes. BMC Evol Biol. 9(1):134.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lowe TM, Chan PP. 2016. TRNAscan-SE On-Line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44 (W1):W54–W57.
- Reineke A, Karlovsky P, Zebitz CPW. 1998. Preparation and purification of DNA from insects for AFLP analysis. Insect Mol Biol. 7(1):95–99.
- Yu X, Tan W, Zhang H, Jiang W, Gao H, Wang W, Liu Y, Wang Y, Tian X. 2019. Characterization of the complete mitochondrial genome of Harpalus sinicus and its implications for phylogenetic analyses. Genes. 10(9):724.