Abstract
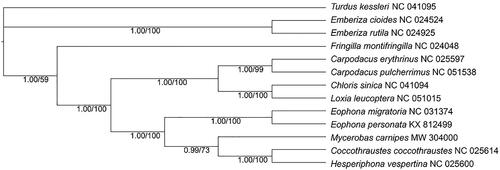
The complete mitochondrial genome of Mycerobas carnipes was sequenced in this study and the total length is 16,806 bp containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and one control region. The phylogenetic analysis based on 13 PCGs of five grosbeaks and other Fringillidae birds demonstrated that Mycerobas, Coccothraustes, and Eophona had close phylogenetic relationships for clustering as three sister branches, and supported that Eophona originated earlier in phylogeny.
Mycerobas carnipes is a grosbeak in the family Fringillidae, whose ecological habitats often occur in forest and shrubland and which distributes in northeastern Iran, the Himalayas to the western Ten-zan, central and southwest China (John and Karen Citation2000). Mycerobas and other grosbeak genera are categorized into the subfamily Coccothraustinae removed from the position of tribe Carduelini (Liang et al. Citation2008). Mycerobas is similar in morphology to Eophona identified by Clement et al. (Citation1993) and the closely relative relationship between the two genera is further confirmed at the molecular level by Liang et al. (Citation2008) based on CoI gene sequence, by Arnaiz-Villena et al. (Citation2001) and Yang et al. (Citation2006) based on Cytb gene sequence. In addition, Eophona is clustered with Coccothraustes in the study of Arnaiz-Villena et al. (Citation2007) based on Cytb gene sequence and of Sun et al. (Citation2016) based on 12 protein-coding genes (PCGs) (except ND6 gene), respectively. However, within the grosbeak genera the phylogenetic relationship is ambiguous. According to Zuccon et al. (Citation2012), the topologies of four grosbeak genera (Coccothraustes, Eophona, Hesperiphona, and Mycerobas) perform differently using the Bayesian inference (BI), the maximum-likelihood (ML) criteria, and different types of datasets. This study presents the complete mitochondrial genome of M. carnipes and constructs a phylogenetic tree based on 13 PCGs of five grosbeaks and other Fringillidae birds for better understanding relationships of grosbeak genera.
The total mitochondrial DNA was extracted from the muscle tissue of M. carnipes, which died of airport protection facility for bird strikes in the Ganzi Gesser Airport, Sichuan Province, China (31.75° N, 99.55° E). The specimen was stored in the Natural Museum of Sichuan University with a voucher number of QZKK091. The complete mitochondrial genome of M. carnipes was sequenced by Chain Termination Method and the genome sequence has been deposited in the GenBank with the accession MW 304000. The assembly of mitochondrial genome was finished via SeqMan software (version 7.1.0), and the annotation was generated by MITOS first (Bernt et al. Citation2013), then corrected manually. The sequence of complete mitogenome is 16,806 bp, including 13 PCGs, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one control region. Sequence analysis showed its total base composition as follows: C (31.4%), A (31.2%), T (23.7%), and G (13.6%); the percentage of A + T (54.9%) was higher than G + C (45.1%), similar to other grosbeaks.
The mitochondrial genome sequence of 13 PCGs of M. carnipes and other 11 Fringillidae species were used for phylogenetic analysis by BI and ML method (). The ML tree was obtained with GTR model with 1000 bootstrap replicates on Mega X (Kumar et al. Citation2018). A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 0.3244)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 31.18% sites). While the BI tree, constructed by PhyloSuite v1.2.2 (Zhang et al. Citation2020) with similar partition model determined by ModelFinder (Kalyaanamoorthy et al. Citation2017) according to AICc, showed completely same topology that the five grosbeaks clustered together sister to Carpodacus, Loxia, and Chloris; and the lineage formed three branches with Eophona basal to them. The phylogenetic result supported that Eophona had earlier origin than other grosbeaks which was different from previous studies with one or a minority of genes (Arnaiz-Villena et al. Citation2007; Zuccon et al. Citation2012). We expect the sequence data will provide a useful data for further study on the phylogenetic evolution of grosbeak genera and other Fringillidae birds.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW 304000.
Additional information
Funding
References
- Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, Lowy E, Zamora J, Varela P, Stefani D, Allende LM. 2001. Phylogeography of crossbills, bullfinches, grosbeaks, and rose finches. Cell Mol Life Sci. 58(8):1159–1166.
- Arnaiz-Villena A, Moscoso J, Ruiz-Del-Valle V, Gonzalez J, Serrano-Vela JI. 2007. Bayesian phylogeny of Fringillinae birds: status of the singular African Oriole Finch (Linurgus olivaceus) and evolution and heterogeneity of genus Carpodacus. Acta Zool Sin. 53:826–834.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Clement P, Harris A, Davies J. 1993. Finches and sparrows: an identification guide. London: Christopher Helm Press.
- John MK, Karen P. 2000. A field guide to the birds of China. Hunan: Education Press.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liang G, Li T, Yin ZH, Lei FM. 2008. Molecular phylogenetic analysis of some Fringillidae species based on mitochondrial CoI gene sequences. Zool Res. 29(5):465–475.
- Sun G, Xia T, Yang X, Zhao C, Liu G, Sha W, Zhang H. 2016. The complete mitochondrial genome sequence of Eophona migratoria (Passeriformes Fringillidae). Mitochondrial DNA Part B. 1(1):753–754.
- Yang SJ, Lei FM, Yin ZH. 2006. Molecular phylogeny of Rose finches and Rose bunting (Passeriformes, Fringillidae, Urocynchramidae). Acta Zootaxon Sin. 31(3):453–458.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zuccon D, Prŷs-Jones R, Rasmussen PC, Ericson PG. 2012. The phylogenetic relationships and generic limits of finches (Fringillidae). Mol Phylogenet Evol. 62(2):581–596.