Abstract
Here, we sequenced and annotated the complete mitochondrial genome (mitogenome) of Palomena viridissima (Hemiptera: Pentatomidae). This mitogenome was 15,118 bp long, comprising of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rrnL and rrnS) and a large non-coding control region. The P. viridissima mitogenome with an A + T content of 76.0%, presented a positive AT-skew (0.11) and a negative GC-skew (-0.13). Ten PCGs started with a typical ATN codon, two PCGs started with TTG (atp8, nad1), whereas the remaining one used AAC (cox1). All tRNAs had a typical secondary cloverleaf structure, except for trnS1 which lacked the dihydrouridine arm. The Bayesian phylogenetic analysis based on mitogenomic data supported a sister relationship of P. viridissima and Nezara viridula from the same tribe Nezarini and recovered a phylogeny of Pentatominae: (Menidini + (Strachiini + (Pentatomini + ((Cappaeini + Halyini) + (Eysarcorini + (Nezarini + Carpocori)))))).
Pentatominae (Insecta: Heteroptera: Pentatomidae), consisting of approximate 4900 species in 938 genera of 40 tribes, is the largest subfamily within Pentatomidae (Eduardo and David Citation2018). Pentatominae species are phytophagous by sucking the sap from the stems, leaves or fruit. To date, phylogenetic analyses within Pentatominae are still limited and mainly employed morphological data (Filipe et al. Citation2017). Here we sequenced and annotated the complete mitochondrial genome (mitogenome) of P. viridissima, which will be helpful to better understand the diversity and phylogeny of Pentatominae.
Adult specimens of P. viridissima were collected from Yuzhong County (104°34′20″E, 36°26′30″N), Gansu Province, China, in July 2018. Samples (voucher number: YZ-7) have been deposited in the State Key Laboratory of Grassland Agro-Ecosystems, College of Pastoral Agricultural Science and Technology, Lanzhou University, Lanzhou, China. The total genomic DNA was extracted from a single specimen using a DNeasy Tissue Kit (Qiagen, German). The P. viridissima mitogenome was amplified by using a set of universal and specific primer pairs, and sequenced in both directions.
The complete mitogenome of P. viridissima was a circular molecule of 15,118 bp long (GenBank accession number: NC_050166), comprising of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA unit genes (rrnL and rrnS) and a large non-coding region (putative control region). The order and orientation of the mitochondrial genes was identical to the inferred ancestral arrangement of insects (Boore Citation1999). Gene overlaps were found at five gene junctions and involved a total of 26 bp, ranging in size from 1 to 8 bp. The longest overlap (8 bp) existed between trnW and trnC. A total of 127 bp intergenic spacers were present in sixteen positions, ranging in size from 1 to 27 bp. The longest intergenic spacers (27 bp) existed between trnS2 and nad1.
The nucleotide composition of the P. viridissima mitogenome was significantly biased toward A and T, with an A + T content of 76.0% (A = 42.3%, C = 13.5%, G = 10.5%, T = 33.7%). This mitogenome presented a positive AT-skew (0.11) and a negative GC-skew (-0.13) on the J-strand. The rrnL was 1,287 bp long with an A + T content of 78.6%, and the rrnS was 803 bp with an A + T content of 77.3%, as found in most insect mitogenomes. Among the 13 PCGs, the lowest A + T content was 69.5% in cox1, while the highest was 82.7% in nad6. Ten PCGs started with a typical ATN codon: two (cox2, nad6) with ATA, three (nad2, nad5 and nad4L) with ATT, four (atp6, cox3, nad4 and cob) with ATG, one (nad3) with ATC. The other two PCGs started with TTG (atp8, nad1) and the remaining one started with AAC (cox1). Nine PCGs terminated with TAA or TAG, whereas four PCGs terminated with an incomplete stop codon T. All of the 22 tRNAs, ranging from 63 bp (trnS1) to 75 bp (trnK), had a typical cloverleaf structure, except for trnS1 which lacked the dihydrouridine arm.
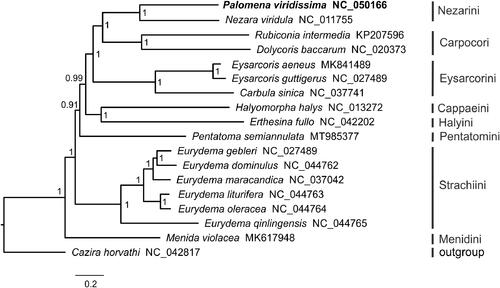
Phylogenetic analysis was performed with the concatenated nucleotide sequences of 13 PCGs and 2 ribosomal RNA genes (rrnS and rrnL) from 17 Pentatominae species and Cazira horvathi (Asopinae, outgroup). We conducted Bayesian inference using MrBayes 3.2.6 (Ronquist and Huelsenbeck Citation2003) on the CIPRES Science Gateway 3.3 (Miller et al. Citation2010). The Bayesian phylogenetic tree supported that P. viridissima clustered with Nezara viridula from the same tribe Nezarini, and recovered a phylogeny of Pentatominae: (Menidini + (Strachiini + (Pentatomini + ((Cappaeini + Halyini) + (Eysarcorini + (Nezarini + Carpocori)))))) ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no NC_050166.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Eduardo IF, David AR, Bolívar RGB 2018. New records of alcaeorrhynchus grandis (Dallas, 1851) (Heteroptera: Pentatomidae) from paraguay, with description of a teratological case. Bol Mus Nac Hist Nat Parag. 22:27–29.
- Filipe MB, Marindia D, Augusto F, Jocelia GV, L. 2017. Total evidence phylogenetic analysis and reclassification of Euschistus Dallas within Carpocorini (Hemiptera: Pentatomidae: Pentatominae). Syst Entomol. 42(2):399–409.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Comp Environ Workshop (GCE). 2010:1–8.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.