Abstract
The freshwater gammarid Grandidierella taihuensis is an important composition of benthic community. In this study, the complete mitogenome sequences of G. taihuensis are determined using next-generation and PacBio long-read sequencing. The mitogenome of G. taihuensis is 15,099 bp in size, and consisted of 13 protein-coding genes, two ribosomal RNA genes, 22 tRNA genes, and a putative control region. Gene arrangement is as same as that of G. rubroantennata. The base composition of the entire mitogenome showed a conspicuous A + T bias of 69.4%. The mitogenome data produced in this study provides a useful resource for future evolutionary and ecological studies.
Introduction
The Grandidierella taihuensis (Morino and Dai Citation1990) is a kind of gammarid. There are more than 40 described species in the genus Grandidierella, and most of them inhabit in the brackish water (Horton et al. 2018). As an important composition of benthic community in freshwater ecosystem, gammarids was used as a water pollution indicator in the Great Lakes in USA (McDonald et al. Citation1990) and Dongting Lake in China (Dai et al. Citation2000), as well as a resource to understand disease epidemiology (Bojko Citation2020). Recently, three complete mitochondrial genome sequences of the genus Grandidierella have been reported (Hiki et al. Citation2020a, Citation2020b). However, G. taihuensis is living in the freshwater (Morino and Dai Citation1990), different from other congeneric brackish-water species. The complete mitochondrial sequences of freshwater gammarid G. taihuensis will help us to understand the phylogeny of Grandidierella in distinct environment. Here, we determined the complete mitochondrial sequences of G. taihuensis using Illumina next-generation sequencing and Pacific Biosciences (PacBio) long read sequencing technology, and performed an analysis of the phylogenetic relationships among Corophioidea species with the available data in GenBank.
Materials and methods
The specimen (NO. TH_CDH09101801; contact person: Jiawen Yin; email: [email protected]) was collected from Changdang Lake in the Taihu Basin on Sep. 10, 2019 (31.624217 N and 119.553693 E), and was stored in herbarium of Freshwater Fisheries Research Institute of Jiangsu Province. Unilateral legs were taken, exoskeleton was removed carefully, and then muscle tissue was gathered for mtDNA isolation. The methods of mtDNA extraction and sequencing were described in our previous work (Zhang et al. Citation2019).
The mitochondria genome was reconstructed basing on both Pacbio Sequel data and the Illumina Hiseq data. After filtered Illumina raw reads, verified the assembly using ABySS (Simpson et al. Citation2009), we completed the circle, filled gaps, and mapped the clean reads to the assembled mitochondria genome, and obtained the complete mitochondrion genome from G. taihuensis. The genes were annotated using MITOS (Bernt et al. Citation2013) and AUGUSTUS (http://bioinf.uni-greifswald.de/augustus/). Transfer RNA (tRNA) genes and Ribosome RNA (rRNA) genes were predicted by tRNAscan-SE (Lowe and Eddy Citation1997) and rRNAmmer version 1.2 (Lagesen et al. Citation2007).
Results and discussion
DNA library of G. taihuensis sample has generated 33,923,743 Illumina paired-end clean reads. The mitogenome of G. taihuensis is 15,099 bp long (GenBank accession numbers: MW118119), with a 30.59% G + C content. A total of 37 genes were predicted, including 13 protein-coding genes (PCGs), two rRNA genes, and 22 tRNA genes. Comparing to pancrustacean ground pattern (Boore Citation1999), there are a translocation of nad6, and a few rearrangement of tRNA genes, just as that of G. rubroantennata (Hiki et al. Citation2020b). Only three protein-coding genes (cob, cox3, and nad3) have ATG as the start codon, four PCGs use ATT (cox1, nad4, nad4L, and nad6), three PCGs use ATA (atp8, cox2, and nad2) and two PCGs use TTG (nad1 and nad5), while atp6 uses ATC. TAA and TAG stop codons are used in 13 genes.
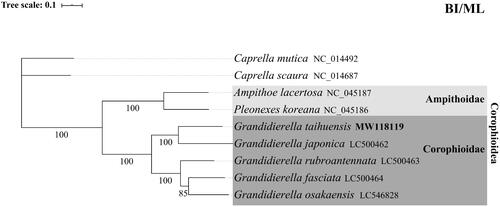
To investigate the phylogenetic relationships of G. taihuensis and other Corophioidea species: Ampithoe lacertosa (NC_045187), Pleonexes koreana (NC_045186), Grandidierella fasciata (LC500464), Grandidierella rubroantennata (LC500463), Grandidierella japonica (LC500462), and Grandidierella osakaensis (LC546828), phylogenetic trees were obtained using maximum likelihood (ML) and Bayesian inference (BI) analyses. Two species in the Caprelloidea were used as outgroup: taxa Caprella mutica (NC_014492) and Caprella scaura (NC_014687). BI and ML trees were performed using MrBayes version 3.2.2 (Ronquist et al. Citation2012) and RAxML (Stamatakis et al. Citation2008), and the GTR model was chosen by jModelTest version 2.1.10 (Darriba et al. Citation2012). The consensus topology of phylogeny trees for Corophioidea species with extremely high supported () show that G. taihuensis and G. japonica is a sister group. Ampithoidae and Corophioidae are monophyly. It is worth noting that a classical mitochondrial barcode fragment (cox1) showed no difference between another freshwater gammarid G. chaohuensis (KT180187), which is also living in China (Hou and Li Citation2002), and our genomic sequences. Further investigation will be needed to reveal the reason of this genetic divergency.
Figure 1. Phylogenetic tree in Corophioidea using protei-coding genes of the complete mitochondrial genome. The complete mitogenome is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood (ML) and Bayesian inference (BI) method. Bootstrap values were presented under the branch, and the posteriori probability values are omitted because all of them are 100%.
Acknowledgments
The authors would like to thank Guosheng Jin for his help in sample collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MW118119. Associated BioProject accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA669588.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bojko J. 2020. The mitochondrial genome of UK (non-native) Dikerogammarus haemobaphes (Amphipoda: Gammaridae) informs upon Dikerogammarus evolution, invasions and associated microparasites. Hydrobiologia. 847(1):229–242.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Dai Y, Tang S, Zhang J. 2000. The distribution of zoobenthos species and bio- assessment of water quality in Dongting Lake. Acta Ecol Sin. 20:277–282.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Hiki K, Ariyama H, Nakajima N. 2020a. The complete mitochondrial genome of the estuarine amphipod Grandidierella osakaensis (Crustacea: Amphipoda). Mitochondrial DNA Part B. 5(3):3323–3324.
- Hiki K, Ariyama H, Nakajima N. 2020b. The complete mitochondrial genomes of two amphipod species of the genus Grandidierella (Crustacea: Amphipoda). Mitochondrial DNA Part B. 5(2):1535–1536.
- Hou Z-E, Li S-Q. 2002. A new species of the genus Grandidierella from Lake Chaohu, China (Crustacea: Amphipoda: Aoridae). Acta Zootaxon Sin. 27:225–234.
- Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Shuangyan Zhao. 2017. Taxonomic study of four species of freshwater Amphipoda(Crustacea: Amphipoda). Heibei University. Master of Science.
- Morino H, Dai A. 1990. Three amphipod species (Crustacea) from East China. Public Itako Hydrobiol Stat. 4:7–27.
- Ronquist F, Teslenko M, Mark Pvd, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6):1117–1123.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771.
- Zhang T, Yin J, Tang S, Li D, Liu X, Gu X, Liu Y. 2019. The complete mitogenome of clam Corbicula fluminea determined usingnext-generation and PacBio sequencing. Mitochondrial DNA Part B. 4(1):1660–1661.
