Abstract
We have assembled and annotated the mitochondrial genome of Tinda javana (Macquart, 1838) (a species of soldier fly) in this study. It is 15,495 bp in length, including 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and a large non-coding control region (length: 704 bp). The nucleotide composition of whole mitochondrial genome biases toward A and T (75.5%). Most PCGs use ATN as initiation codon, except for cox1 which starts with CGA. All PCGs end with common termination codon TAA/G. Phylogenetic analyses based on the nucleotide sequence data supported the monophyly of Stratiomyidae and the sister relationship between Pachygastrinae and the clade (Nemotelinae + (Hermetiinae + Sarginae)).
Pachygastrinae is a subfamily of flies, which belongs to the family Stratiomyidae of Diptera. It is a larger subfamily in Stratiomyidae and widely distributes all over the world. Up to now, 619 species of 180 genera are known worldwide (Woodley Citation2001; Yang, Zhang, et al. Citation2014). Larvae of some species feed on Musa L. plants and are important agricultural and forestry pests (Yang ZH, Yang Y, et al. Citation2014). Classification and systematics of Pachygastrinae are of great interest for management of plant pests. This study firstly sequenced the mitochondrial genome of Tinda javana to discuss the phylogenetic status of Pachygastrinae.
The samples of T. javana were collected from Luodian County (E106.7917, N25.4075), Guizhou, China, and deposited in Insect Herbarium of Guizhou Academy of Forestry, Guiyang (GZAF-2020-DS1445) (URL, Zaihua Yang and [email protected]). The total genome DNA was sequenced using Illumina MiSeq format (Illumina, San Diego, CA). Then the data was assembled by NOVOPlasty version 2.7.0 (Dierckxsens et al. Citation2017) with the cox1 gene of Nemotelus notatus (MT584142) as the seed. The complete mitochondrial genome was annotated using MITOZ version 1.04 (Meng et al. Citation2019). All 37 mitochondrial gene sequences were aligned using MAFFT version 7.394 (Kuraku et al. Citation2013) with G-INS-I (accurate) strategy. Maximum likelihood (ML) tree was inferred by IQ-TREE version 1.6.3 (Nguyen et al. Citation2015) under the optimal model (GTR + I + G for Subset1 (nad3, atp6, cox1, cox2, cytb, and cox3), Subset2 (atp8, nad2, and nad6), and Subset3 (nad1, nad4L, nad4, and nad5); TVM + G for Subset4 (rrnL and rrnS); TVM + I + G for Subset5 (trnM, trnW, trnK, trnV, trnC, trnI, trnP, trnH, trnE, trnD, trnT, trnL1, trnL2, trnN, trnG, trnS2, trnR, trnA, and trnS1); HKY + G for Subset6 (trnQ, trnY, and trnF)).
The complete mitogenome of T. javana (Genbank accession no. MW115422) is 15,495 bp in length with highly AT biased of nucleotide composition (38.1% A, 37.4% T, 9.9% G, and 14.6% C). All 37 typical mitochondrial genes were annotated in this newly sequenced species, containing 13 PCGs, 22 tRNAs, two rRNAs, and control region. The mitochondrial genome of T. javana shares the same gene order as ancestral insects such as Drosophila yakuba and Drosophila mercatorum (Clary and Wolstenholme Citation1985; Wang et al. Citation2019). Fifteen tRNAs and nine PCGs are encoded on the majority strand (J-strand) while the remaining genes are encoded on the minority strand (N-strand). A total of ten overlaps (1–8 bp) between adjacent genes were found, the longest overlap is 8 bp between trnW and trnC. The intergenic spacers were detected in 12 locations, ranging from 1 to 18 bp. The length of 13 PCGs is 11,154 bp, accounting for 72.06% of whole mitochondrial genome. Most PCGs initiated by standard start codon ATN (ATA/T/G/C), except for cox1 which starts with CGA. All PCGs end with common termination codon TAA/G.
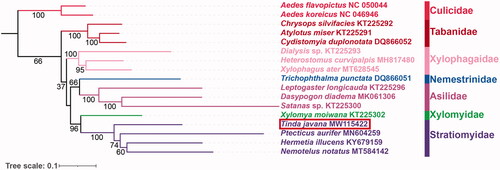
Here, based on the nucleotide sequence data of 37 genes from T. javana and other 16 species belonging to seven related families of Diptera, we reconstructed the phylogenetic tree (). The ML analyses strongly supported the monophyly of Stratiomyidae (BS = 100), consistent with some previous studies (Zhou et al. Citation2017; Ding and Yang Citation2020). In Stratiomyidae, the relationships among included subfamilies are inferred as (Pachygastrinae (T. javana) + (Nemotelinae (Nemotelus notatus) + (Hermetiinae (Hermetia illucens) + Sarginae (Ptecticus aurifer)))).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW115422. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA705515, SRR13810270, and SAMN18091015, respectively.
Additional information
Funding
References
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Ding SM, Yang D. 2020. Complete mitochondrial genome of Coenomyia ferruginea (Scopoli). (Diptera, Coenomyiidae). Mitochondrial DNA B Resour. 5(3):2711–2712.
- Kuraku S, Zmasek CM, Nishimura O, Katoh K. 2013. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41(Web Server issue):W22–W28.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wang AT, Du ZY, Luo X, Zhang FC, Zhang JZ, Li H. 2019. The conserved mitochondrial genomes of Drosophila mercatorum (Diptera: Drosophilidae) with different reproductive modes and phylogenetic implications. Int J Biol Macromol. 138:912–918.
- Woodley NE. 2001. A world catalog of the Stratiomyidae (Insecta: Diptera). Myia. 11:1–475.
- Yang D, Zhang TT, Li Z. 2014. Stratiomyoidea of China. Beijing, China: China Agricultural University Press; p. 1–870.
- Yang ZH, Yang Y, Yang MF. 2014. Morphological characteristics and control of Parastratiosphecomyia szechuanensis, a new Musa pest. Guizhou Agric Sci. 42(9):134–136.
- Zhou QX, Ding SM, Li X, Zhang TT, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA B Resour. 2(2):461–462.