Abstract
In this study, the complete chloroplast genome sequence of Pterocarya macroptera var. delavayi was reported and characterized. The chloroplast genome is 160,168 bp in length, and consists the typical quadripartite structure, a pair of inverted repeats (IRs, 26,007 bp) separated by a large single-copy region (89,701 bp) and a small single-copy region (18,453 bp). A total of 136 unique genes were predicted, including 88 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. The GC content of the chloroplast genome is 36.2%. Phylogenetic analysis confirmed the close relationship between Pterocarya and Juglans.
Pterocarya macroptera var. delavayi distributes in western Hubei, western Sichuan, and northwest Yunnan of China (Lu et al. Citation1999). It is a riparian dominant tree, and its bark can be used as fiber raw material (Lu et al. Citation1999; Ying and Chen Citation2011). P. macroptera var. delavayi is a variety of P. macroptera, which is morphologically different from P. macroptera var. macroptera by its mature leaves with exclusively solitary trichomes (Song et al. Citation2020). Based on RAD-seq data, Song et al. (Citation2020) showed that the genus Pterocarya is monophyletic and close to Juglans. P. macroptera var. delavayi was sister to the other two varieties (P. macroptera var. macroptera and P. macroptera var. insignis) (Song et al. Citation2020).
Here, the complete chloroplast genome of P. macroptera var. delavayi was determined, annotated, and analyzed. The sample was collected from the Botanical Garden Edinburgh (UK) (55°56′54.56″N, 3°1′57.59″ W) by Jianfei Ye. The DNA sample (RBGE 2019-08-27) was deposited in the Institute of Life Science, Nanchang University (JXU). Total genomic DNA was extracted from silica gel-dried leaves using the modified CTAB method (Doyle Citation1987) and sequenced using MGI MGISEQ-2000 High-throughput Sequencing Set. In total, 3 Gb of 150-bp paired-end raw reads were generated and used for chloroplast genome assembly. Trimmomatic version 0.39 (Bolger et al. Citation2014) was used to organize and trim overrepresented sequences for getting the clean reads. The clean reads were assembled by using GetOrganelle version 1.5 (Jin et al. Citation2020). The chloroplast genome of P. macroptera var. delavayi was annotated used Geneious version 9.05 (http://www.geneious.com/) with P. stenoptera (NC_046428) as reference. The annotated complete chloroplast genome of P. macroptera var. delavayi was deposited in GenBank (the accession number MW194257).
The complete chloroplast genome of P. macroptera var. delavayi is 160,168 bp in length, with a large single-copy region (LSC) of 89,701 bp, a small single-copy region (SSC) of 18,453 bp, and a pair of inverted repeat regions (IRs) of 26,007 bp, and forms a typical quadripartite structure. A total of 136 genes were annotated from the chloroplast genome of P. macroptera var. delavayi, including 88 protein-coding genes, 40 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The GC content of the chloroplast genome is 36.2%, meanwhile, the corresponding value of LSC, SSC, and IR regions is 33.8%, 29.8%, and 42.6%, respectively.
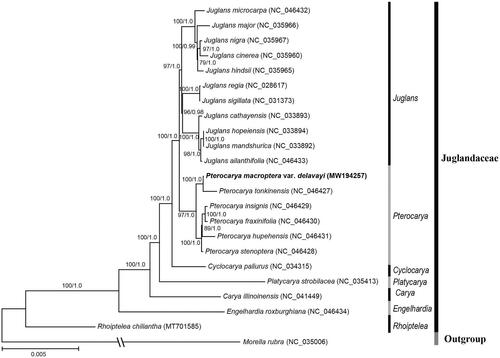
In order to analyze the phylogenetic position of P. macroptera var. delavayi, another 22 complete chloroplast genomes were retrieved from NCBI, and Morella rubra from Myricaceae was set as the outgroup (). All sequences were aligned by MAFFT version 7.409 (Katoh and Toh Citation2010). The maximum likelihood tree and Bayesian tree were constructed using RAxML version 8.2.12 (Stamatakis Citation2014) and MrBayes version 3.2.6 (Ronquist and Huelsenbeck Citation2003), respectively. The phylogenetic tree showed that Pterocarya is formed a clade with the Juglans with high support (ML-PP = 100%, BI-PP = 1.0), and P. macroptera var. delavayi is sister to P. tonkinensis with strongly supported values (ML-PP = 100%, BI-PP = 1.0, ). The phylogenetic relationships among Juglandaceae were consistent with the results of Xiang et al. (Citation2014) and Mu et al. (Citation2020) based on chloroplast genome data.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no.MW194257. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA692030, SRR13434420, and SAMN17304756, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Lu AM, Stone DE, Grauke LJ. 1999. Juglandaceae. In: Wu ZY, Raven PH, editors. Flora of China, Vol. 4. Beijing, China: Science Press & Missouri Botanical Garden Press; p. 277–285.
- Mu XY, Tong L, Sun M, Zhu YX, Wen J, Lin QW, Liu B. 2020. Phylogeny and divergence time estimation of the walnut family (Juglandaceae) based on nuclear RAD-Seq and chloroplast genome data. Mol Phylogenet Evol. 147:106802.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Song YG, Li Y, Meng HH, Fragnière Y, Ge BJ, Sakio H, Yousefzadeh H, Bétrisey S, Kozlowski G. 2020. Phylogeny, taxonomy, and biogeography of Pterocarya (Juglandaceae). Plants. 9(11):1524.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xiang XG, Wang W, Li RQ, Lin L, Liu Y, Zhou ZK, Li ZY, Chen ZD. 2014. Large-scale phylogenetic analyses reveal fagalean diversification promoted by the interplay of diaspores and environments in the Paleogene. Perspect Plant Ecol. 16(3):101–110.
- Ying TS, Chen ML. 2011. Plant geography of China. Shanghai, China: Shanghai Scientific & Technical Publishers; p. 141.