Abstract
In this study, we assembled the complete mitochondrial genome of Yanglong yak (Bos grunniens) from Illumina sequencing reads. The mitochondrial genome is 16,323 bp long with an A + T-biased nucleotide composition, and encodes 13 protein-coding, 22 tRNA, and two rRNA genes along with a noncoding control region. In addition, its gene order is identical to those of the previously published mitochondrial genomes of its congeners. Phylogenetic analysis indicates that this breed is closely related to Datong yak, Pamir yak, Tianjun yak, polled yak, Seron yak, Sunnan yak, a series of Domestic Yak and wild yak, followed by Jinchuan yak and Gannan yak, and slightly far away from Bison and Bos taurus.
The genetic characteristics of the mitochondrial genome are one of the important means to study molecular biology (Lorenzo et al. Citation2016). The animal mitochondrial genome is characterized by simple structure and easy detection. Moreover, as the mitochondrial genome is inherited from the mother line, it is very beneficial to study the genetic evolution and phylogeny of the population. For centuries, yaks have been maintained by local nomadic pastoralists for transportation, milk, butter, meat, and other byproducts (Leslie and Schaller Citation2009; Qiu et al. Citation2012). Bos grunniens and Bos mutus are domestic and wild forms of yak, respectively, and are both native to the Qinghai-Tibetan Plateau and adjacent high-altitude regions (Shi et al. Citation2016; Chen et al. Citation2018). Yanglong yak is a new breed bred from Qinghai Plateau Yak and wild yak after long-term natural breeding and artificial mating (Shi et al. Citation2016). It has been living in cold and hypoxia environment for many years, which has the characteristics of rough feeding tolerance and environmental adaptability. Therefore, this study assembled the complete mitochondrial genome for Yanglong yak from Qinghai Province of China, and investigated its genetic relatedness to other taxa within the subfamily Bovinae (Cetartiodactyla: Bovidae).
The sample selected for this study was a 3-year-old healthy female yak, and the blood sample was collected from Yanglong Township, Qilian County, Haibei Tibetan Autonomous Prefecture, Qinghai Province (38.69°N, 98.55°E), and was used for DNA isolation with the QIAamp DNA Blood Mini Kit (Qiagen, CA, USA). A specimen was deposited at the Key Laboratory of Yak Breeding Engineering of Gansu Province, Lanzhou Institute of Husbandry and Pharmaceutical Sciences (Lanzhou, Gansu Province, China, Xian Guo and [email protected]) under the voucher number BMXBG2020053. The genomic DNA was extracted from Yanglong yak, is stored at −80 °C (ultra-deep-freeze refrigerator) in the sample storage room of our department. Library construction and paired-end DNA sequencing with Illumina HiSeq X™ Ten Sequencing System (Illumina, CA, USA) were conducted by Annoroad Gene Technology (Beijing, China), which yielded a total of 15.33 M 150-bp raw paired reads. The program MITObim version 1.9 (Hahn et al. Citation2013) was employed to assemble the mitochondrial genome along with a previously published reference sequence (GenBank accession: JQ692071) (Qiu et al. Citation2012). The mitogenome annotation was done by comparing with those of its congeners.
The mitochondrial genome of the Yanglong yak (GenBank accession: MT649466) was successfully assembled with an average coverage of 27.1X. It is 16,323 bp long with an A + T-biased nucleotide composition (33.7% A, 25.8% C, 13.2% G, and 27.3% T; ‘light strand’), and encodes the typical set of 37 animal mitochondrial genes (incl. 13 protein-coding, 22 tRNA, and 2 rRNA genes) along with a noncoding control region. Its gene order and codon usage are highly conserved, and are identical to those of the previously published mitochondrial genomes of its congeners (e.g. Hiendleder et al. Citation2008; Wu et al. Citation2016; Guo et al. Citation2019). In all, the 13 protein-coding genes involve two types of initiation codons (ATA & ATG) and three types of termination codons (TAA, TAG & T). The 22 tRNAs range in size from 60 (tRNA-SerAGY) to 75 bp (tRNA-LeuUUR) with a total length of 1509 bp. The secondary structures of the tRNA genes are analyzed and predicted by tRNAscan-SE version 2.0 online website, which is similar to that of other mammals, except that tRNA-SerAGY cannot form the typical clover structure due to the lack of the DHU arm. In addition to tRNA-His, tRNA-Ile, tRNA-Lys, and tRNA-Tyr, a total of 43 base pair mismatches (34 G-U, 3 A-C, 3 U-U, 2 A-G, and 1 C-U) were found by the prediction analysis of 22 tRNA pairs, about 80% of which are G-U mismatches. The tRNA-UCN is the most base pair mismatched gene with 5 G-U mismatches. The two rRNAs are 957 bp (12S rRNA) and 1571 bp (16S rRNA) long, respectively. The control region is 893 bp long with an A + T-biased nucleotide composition (60.9% A + T; ‘light-strand’). Besides, a 31-bp-long origin of L-strand replication is present between tRNA-Asn and tRNA-Cys.
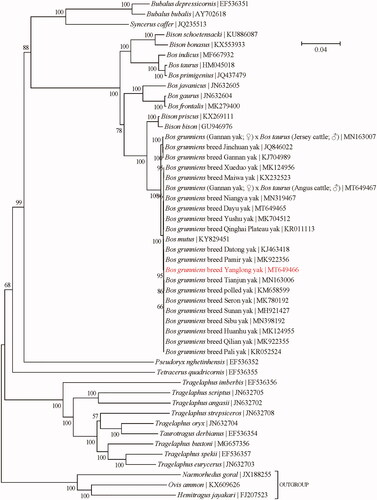
To investigate the genetic relatedness of Yanglong yak to the other taxa within the subfamily Bovinae, a maximum-likelihood (ML) phylogenetic analysis was conducted using the concatenated sequences of all 13 protein-coding genes (alignment size: 11,370 bp) with the program PhyML-aLRT version 2.4.5 (Guindon and Gascuel Citation2003) as implemented in TOPALi version 2.5 (Milne et al. Citation2009) (). The best-fit nucleotide substitution model is ‘GTR + G+I’. Bootstrap support values were estimated from 100 random runs following the method of Anisimova and Gascuel (Citation2006). The outgroup taxa employed in the phylogenetic analysis are three confamilial species from the subfamily Caprinae, including Hemitragus jayakari (FJ207523) (Hassanin and Douzery Citation1999), Naemorhedus goral (JX188255) (Yang et al. Citation2013), and Ovis ammon (KX609626) (Mao et al. Citation2017). The yak in this study was found to be closely related to Datong yak, Pamir yak, Tianjun yak, polled yak, Seron yak, Sunnan yak, a series of Domestic Yak and wild yak, followed by Jinchuan yak and Gannan yak, and slightly far away from Bison and Bos taurus. The high genetic similarity between domestic and wild yaks was also previously confirmed by Zhang et al. (Citation2009). In addition, it also revealed that the two genera Bison and Bos failed to form two distinct monophyletic groups, which could be attributed to the frequent hybridization between them (Douglas et al. Citation2011; Shi et al. Citation2016).
Figure 1. Phylogeny of the subfamily Bovinae based on the maximum-likelihood analysis of the concatenated sequences of 13 mitochondrial protein-coding genes (alignment size: 11,370 bp). The best-fit nucleotide substitution model is ‘GTR + G+I.’ The bootstrap support values next to the nodes are based on 100 random runs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MT649466.
Additional information
Funding
References
- Douglas KC, Halbert ND, Kolenda C, Childers C, Hunter DL, Derr JN. 2011. Complete mitochondrial DNA sequence analysis of Bison bison and bison-cattle hybrids: function and phylogeny. Mitochondrion. 11(1):166–175.
- Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 55(4):539–552.
- Chen M, Sun Y, Yang C, Zeng G, Li Z, Zhang J. 2018. The road to wild yak protection in China. Science. 360(6391):866.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704.
- Guo X, Wu X, Bao P, Zhao S, Ma J, Yang S, Chu M, Ding X, Yao X, Liang C, et al. 2019. Characterization of the complete mitochondrial genome of the Tianjun yak (Bos grunniens). Mitochondrial DNA Part B. 4(2):4120–4121.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- Hassanin A, Douzery EJP. 1999. The tribal radiation of the family Bovidae (Artiodactyla) and the evolution of the mitochondrial cytochrome b gene. Mol Phylogenet Evol. 13(2):227–243.
- Hiendleder S, Lewalski H, Janke A. 2008. Complete mitochondrial genomes of Bos taurus and Bos indicus provide new insights into intra-species variation, taxonomy and domestication. Cytogenet Genome Res. 120(1–2):150–156.
- Leslie DM, Jr., Schaller GB. 2009. Bos grunniens and Bos mutus (Artiodactyla: Bovidae). Mamm Species. 836:1–17.
- Lorenzo PD, Lancioni H, Ceccobelli S, Curcio L, Panella F, Lasagna E. 2016. Uniparental genetic systems: a male and a female perspective in the domestic cattle origin and evolution. Electron J Biotechn. 23:69–78.
- Mao H, Liu H, Ma G, Yang Q, Guo X, Lamaocao Z. 2017. The complete mitochondrial genome of Ovis ammon darwini (Artiodactyla: Bovidae). Conservation Genet Resour. 9(1):59–62.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127.
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44(8):946–949.
- Shi Q, Guo Y, Engelhardt SC, Weladji RB, Zhou Y, Long M, Meng X. 2016. Endangered wild yak (Bos grunniens) in the Tibetan plateau and adjacent regions: Population size, distribution, conservation perspectives and its relation to the domestic subspecies. J Nat Conserv. 32:35–43.
- Wu X, Ding X, Chu M, Guo X, Bao P, Liang C, Yan P. 2016. Characterization of the complete mitochondrial genome sequence of Gannan yak (Bos grunniens). Mitochondrial DNA Part A 27(2):1014–1015.
- Yang C, Xiang C, Qi W, Xia S, Tu F, Zhang X, Moermond T, Yue B. 2013. Phylogenetic analyses and improved resolution of the family Bovidae based on complete mitochondrial genomes. Biochem Syst Ecol. 48:136–143.
- Zhang ZG, Xia L, Yang QS. 2009. Distribution and conservation of yak (Bos grunniens). Chin J Zool. 44(1):148–150.
