Abstract
The greater argonaut Argonauta argo is a species of the paper nautilus (Argonautidae), which is a family in Octopoda. In this paper, we report its full mitogenome sequence, which was obtained from a specimen collected in the Japan Seas near Oki Island, Shimane Prefecture, in Japan. The sequence was determined using the NGS Illumina HiSeq platform. With its 37 genes, the mitogenome shows a typical metazoan and Octopoda genomic structure, and similar to the mitogenome of the previously reported congener, A. hians. To confirm A. argo phylogenetic position in Octopoda, we conducted maximum likelihood phylogenetic analysis, using a data set including publicly available 17 Octopodiformes, five Decapodiformes, three Nautiloids and two outgroup Conchiferans. The result confirmed the affinity of Argonautidae to Tremoctopus, and the sister group position of this clade against the rest of incirrate Octopods. The mitogenome and phylogeny of A. argo reported here will be useful for future studies involving this enigmatic species, including on the reacquisition of external calcified shell structures in mollusks.
The greater paper nautilus Argonauta argo is the largest species of the sole extant genus of the pelagic octopod Argonautidae (Young et al. Citation1998). Similar to its congener, A. hians, it is a cosmopolitan species inhabiting the tropical and subtropical open seas worldwide (Norman Citation2000). Also similar to all females of the genus, female A. argo produces a thin, brittle calcite egg cases formed by two specialized dorsal arms, which outer form resembles the shape of Nautiloids’ and Ammonoids’ shells (Scales Citation2015; Stevens et al. Citation2015). This ‘shell’ (egg case) is thought not to be a homologous structure to the calcified shells of Conchiferans and Cephalopods, but an evolutionary innovation (an apomorphy) of the genus (Naef Citation1923).
In this paper, we report the full mitochondrial genome (mitogenome) sequence of A. argo. A piece of the gonad (ovary) was collected from an individual female specimen and was stored frozen at −80 °C until DNA extraction. The individual was collected in 2018 from the Sea of Japan around Oki Island (36°17′20.6″N, 133°12′46.4″E) of Shimane Prefecture in Japan. The shell is registered as a collection of The University Museum, The University of Tokyo in Tokyo, Japan (Voucher No. RM33391). Genomic DNA was extracted from the ovary using the QIAGEN Genomic-tip kit. The DNA sample was then analyzed on a HiSeq Illumina Next Generation Sequencer. To reconstruct the full mitochondrial genome, we performed contig assembly (-n 200) with Platanus v1.2.4 (Kajitani et al. Citation2014) using the paired-end data. Contigs annotated as mitochondrial sequences were extracted by using the mitogenome data of a closely related species, A. hians (NC_036354), as the query for BLASTn homology search. After assembling the contigs, both ends of the resulting single contig were manually confirmed to overlap, and redundant parts were removed to complete the full circular mitogenome. Afterward, the full mitogenome sequence was annotated using the web server version of MITOS (Bernt et al. Citation2013).
Argonauta argo’s mitogenome sequence was 15,741 bases-long (registered to DDBJ; Accession Number LC596061). Its structure is identical to the previously reported mitogenome of Argonauta hians (Chiu et al. Citation2018) and to those of the order Octopoda in general (Cheng et al. Citation2013; Magallón-Gayón et al. Citation2020). Detailed genomic structure is as follows: (1) There are 13 protein-coding, two rRNA, and 22 tRNA genes; (2) One control region is located between the COX3 and tRNA-Glu (817 bp-long); (3) Seven of the 13 protein-coding genes are coded on the L chain (ATP6, ATP8, COII, COI, ND2, ND3, and COIII); (4) ND4L is located in tandem with ND4 on the H chain; (5) Both the SSU-rRNA and LSU-rRNA coding genes are located on the H chain; (6) eight tRNA genes (tRNA-Ser(agc), tRNA-Thr, tRNA-Ile, tRNA-Asn, tRNA-Arg, tRNA-Ala, tRNA-Lys, and tRNA-Asp) are located on the L chain. The total GC content of the mitogenome was 23.3%, similar to other members of Octopoda (Cheng et al. Citation2013; Magallón-Gayón et al. Citation2020). To check the phylogenetic position of A. argo among the octopods, we collected mitogenome sequences of one polyplacophoran and one gastropod as outgroups, three nautiloids, five decapodiforms, and 17 octopodiforms from GenBank, and included them in the data set for phylogenetic analyses. A maximum likelihood phylogenetic analysis (Yang Citation1994) using the GUI version of RAxML (Silvestro and Michalak Citation2012; Stamatakis Citation2014) was conducted. Detailed methods of the analysis are shown in the legend of .
Figure 1. The maximum likelihood (ML) phylogenetic tree showing the position of Argonauta argo. The support values of nodes with 100% bootstrap supports are not shown. Phylogenetic analyses were conducted on a data matrix (10,428 positions) including all concatenated nucleotide sequences of the mitogenomes except the third codon positions of the protein-coding genes. Gene sequences were aligned individually using the online version of MAFFT under default settings (Katoh and Standley Citation2013). Aligned sequences were individually edited using the online version of GBlocks using the least stringent settings (Castresana Citation2000). A partitioned ML analysis (three partitions: protein-coding, rRNA, tRNA) were performed using the RAxML-GUI ver. 1-5b1 (Silvestro and Michalak Citation2012; Stamatakis Citation2014), with the GTR + Γ nucleotide substitution model (Yang Citation1994) 1000 rapid bootstrap replications.
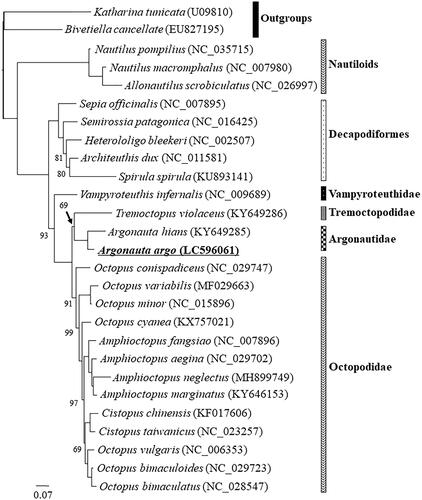
The topology of the obtained ML tree is shown in . The resulting phylogeny showed that A. argo formed a group to its congener, A. hians, forming a monophyletic Argonautidae, with the blanket octopus Tremoctopus as its sister group (superfamily Argonautoidea) () (Strugnell et al. Citation2006; Sanchez et al. Citation2018). Our phylogenetic tree also showed that Argonautoidea is sister to the rest of incirrate Octopoda (Uribe and Zardoya Citation2017; Chiu et al. Citation2018). We are confident that the result presented here will be useful for future molecular phylogenetics studies addressing taxonomic and systematic questions in Argonautidae, Argonautoidea, and Octopoda, beside for future phylogeography and population genetics studies of this species.
Acknowledgments
The authors would like to thank Masaki Yamamoto (National Institute of Technology (KOSEN), Wakayama College), Takenori Sasaki and Nagisa Nakaji (The University Museum, The University of Tokyo) for their assistance on sampling, molecular experiments, and data-set building for the phylogenetic analyses in this study. Genome sequencing was done with the support of the Platform for Advanced Genome Science (PAGS) Project.
Disclosure statement
The authors declare that there is no conflict of interest. All authors are responsible for the content and writing of the paper.
Data availability statement
The full mitochondrial genome sequence reported in this study is registered in and openly available from the National Center for Biotechnology Information (NCBI) Genbank database (Accession No.: LC596061; https://www.ncbi.nlm.nih.gov/nuccore/LC596061.1).
The shell (eggcase) of the specimen was deposited at The University Museum of The University of Tokyo, Japan (Takenori Sasaki; [email protected]) under the voucher number UMUT-RM33391.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Cheng R, Zheng X, Ma Y, Li Q. 2013. The complete mitochondrial genomes of two octopods Cistopus chinensis and Cistopus taiwanicus: revealing the phylogenetic position of the genus Cistopus within the Order Octopoda. PLoS One. 8(12):e84216.
- Chiu YW, Chang CW, Lin HD, Shen KN. 2018. The complete mitogenome of the winged argonaut Argonauta hians and its phylogenetic relationships in Octopoda. Conservation Genet Resour. 10(3):359–362.
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Magallón-Gayón E, del Río-Portilla MÁ, de los Angeles Barriga-Sosa I. 2020. The complete mitochondrial genomes of two octopods of the eastern Pacific Ocean: Octopus mimus and 'Octopus' fitchi (Cephalopoda: Octopodidae) and their phylogenetic position within Octopoda. Mol Biol Rep. 47(2):943–952.
- Naef A. 1923. Die cephalopoden. Fauna et Flora Del Golfo di Napoli. 35:1–148.
- Norman M. 2000. Cephalopods: a world guide. Hackenheim: ConchBooks; p. 189–195.
- Sanchez G, Setiamarga DHE, Tuanapaya S, Tongtherm K, Winkelmann IE, Schmidbaur H, Umino T, Albertin C, Allcock L, Perales-Raya C, et al. 2018. Genus-level phylogeny of cephalopods using molecular markers: current status and problematic areas. PeerJ. 6:e4331.
- Scales H. 2015. Spirals in time: the secret life and curious afterlife of seashells. London: Bloomsbury Publishing.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Stevens K, Iba Y, Suzuki A, Mutterlose J. 2015. Biological and environmental signals recorded in shells of Argonauta argo (Cephalopoda, Octobrachia) from the Sea of Japan. Mar Biol. 162(11):2203–2215.
- Strugnell J, Jackson J, Drummond AJ, Cooper A. 2006. Divergence time estimates for major cephalopod groups: evidence from multiple genes. Cladistics. 22(1):89–96.
- Uribe JE, Zardoya R. 2017. Revisiting the phylogeny of Cephalopoda using complete mitochondrial genomes. J Mollusc Stud. 83(2):133–144.
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 39(3):306–314.
- Young RE, Vecchione M, Donovan DT. 1998. The evolution of coleoid cephalopods and their present biodiversity and ecology. S Afr J Mar Sci. 20(1):393–420.
