Abstract
Thalassiosira nordenskioeldii is a common harmful algal bloom (HAB) species with worldwide distribution. Although barcode sequences of this ecologically important species have been published, no genome data have been published for T. nordenskioeldii. In this study, we constructed the complete mitochondrial genome (mtDNA) of a T. nordenskioeldii strain isolated from the East China Sea. The T. nordenskioeldii mtDNA is circular and has a length of 47,038 bp and a GC content of 30.84%. The mtDNA encodes 69 genes, including 40 protein-coding (PCGs), 27 tRNA and two rRNA genes. Phylogenetic analysis using concatenated amino acid sequences of 31 shared PCGs from 37 diatom mtDNAs revealed that the mtDNA of T. nordenskioeldii was fully resolved in a clade with that of Thalassiosira profunda. The mtDNA of T. nordenskioeldii showed high collinearity with those of T. profunda and Skeletonema marinoi with only minor rearrangements. The completion of Thalassiosira mtDNAs will facilitate evolutionary studies on species of the order Thalassiosirales and the class Mediophyceae.
The type species of the genus Thalassiosira (Cleve Citation1873), Thalassiosira nordenskioeldii Cleve Citation1873, is a harmful algal bloom (HAB) species with worldwide distribution (Hoppenrath et al. Citation2007; Li et al. Citation2013). Although T. nordenskioeldii is a common HAB species that frequently causes HABs, molecular information of this species is still limited. Sequences of only a few common molecular markers of T. nordenskioeldii including cox1, rbcL and 18S rDNA have been published (Eharam, Inagaki, et al. Citation2000a; Ehara, Watanabe, et al. 2000; Alverson et al. Citation2007). Notably, a group II intron has been characterized in the cox1 gene of T. nordenskioeldii (Ehara, Watanabe, et al. Citation2000b). Here, we constructed the first complete mtDNA of T. nordenskioeldii using the strain CNS00052 isolated from water samples collected in the East China Sea (33°59.934′N, 121°13.59′E) in April 2019. Its specimen is deposited in the collection of marine algae in KLMEES of IOCAS (Nansheng Chen, [email protected]) under the voucher number CNS00052.
The total DNA was extracted using DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). The DNA Library was prepared using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA). After qualification, the library was sequenced using an Illumina NovaSeq 6000 platform (Novogene, Beijing). Sequencing results were assembled using Platanus Allee v2.2.2 (Kajitani et al. Citation2019), and the target scaffolds were selected using BLASTN v2.10.0. BWA v0.7.17 (Li and Durbin Citation2010) was used to align and verify the circular sequence of the mtDNA. The complete T. nordenskioeldii mtDNA (GenBank accession number: MW387419) was 47,038 bp in length with a GC content of 30.84%. The circular mtDNA encodes 69 genes, including 27 tRNA genes, two rRNA genes and 40 protein-coding genes (PCGs), which include five open reading frames (orfs) with unknown functions. Introns have been found to occur frequently in cox1 of T. nordenskioeldii strains, such as the CCMP992 strain (AB038235) isolated from the east coast of USA and the CCMP997 strain from Norway (Ehara, Watanabe, et al. 2000). However, no intron was found in the complete cox1 of the CNS00052 strain, whose cox1 was 1,521 bp in length encoding 506 amino acids, suggesting high genomic variations among T. nordenskioeldii strains. Comparative analysis of the full-length cox1 genes of T. nordenskioeldii strains indicated that the cox1 genes (AB038235 and AB037974) reported previously (Ehara, Watanabe, et al. 2000) were incomplete. Of the 40 PCGs, 37 initiated with the ATG start codon, while the other three PCGs started with TTG, GTG and ATT, respectively. The stop codons of the 40 PCGs included TAA (31 of 40) and TAG (9 of 40).
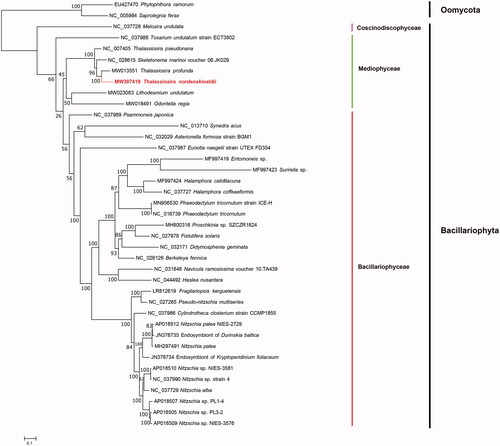
To construct the maximum likelihood (ML) phylogenetic tree, all of the 31 PCGs, including atp6, 8, 9; cob; cox1, 2, 3; nad1-7, 4 L, 9, 11; rpl2, 5, 6, 14, 16; rps3, 4, 8, 10, 11, 13, 14, 19; and tatC from 37 diatom mtDNAs including the mtDNA of T. nordenskioeldii, and the mtDNAs of two outgroup taxa, Oomycota species Phytophthora ramorum (EU427470) and Saprolegnia ferax (NC_005984), were first aligned using MAFFT v7.471 (Katoh and Standley Citation2013) and trimmed using trimAL v1.4 (Capella-Gutierrez et al. Citation2009) individually with default parameters (). Amino acid sequences of all 31 PCGs of 39 species (including T. nordenskioeldii) were concatenated using Phyutility (Smith and Dunn Citation2008), and the best-fit models for all PCGs were predicted using ModelFinder (Kalyaanamoorthy et al. Citation2017) with four categories for each partition and a bootstrap value of 1000. The phylogenetic tree was constructed using IQtree v1.6.12 (Trifinopoulos et al. Citation2016). In the phylogenetic tree, Thalassiosira nordenskioeldii grouped with species from Thalassiosira and Skeletonema species with 100% support, which belong to the class Mediophyceae, order Thalassiosirales (Stoermer Citation2003). Thalassiosira nordenskioeldii clustered well with T. profunda (MW013551). The tree topologies was similar to those reported previously by Kaczmarska et al. (Citation2006) and Alverson et al. (Citation2007), which were constructed using the full-length 18S rDNA, the partial 28S rDNA and two chloroplast DNA (psbC and rbcL) sequences. All phylogenetic trees showed that T. nordenskioeldii and T. profunda of the family Thalassiosiraceae had closer evolutionary relationship with Skeletonema marinoi of the family Skeletonemataceae than that with T. pseudonana, although Thalassiosiraceae and Skeletonemataceae were different families in the order Thalassiosirales (Stoermer Citation2003), supporting the proposal that the taxonomical position of T. pseudonana needs to be revised (Alverson et al. Citation2011). Furthermore, the mtDNA of T. nordenskioeldii showed high collinearity with those of T. profunda and S. marinoi (NC_028615) with only minor rearrangements. Two translocation events situated atp6 and rps10 to the region between rns and rnl, and another translocation event brought atp9 to the position between nad2 and nad6. The completion of more mtDNAs of Thalassiosira species is critical for comparing and classifying species of the order Thalassiosirales and the class Mediophyceae.
Figure 1. Maximum likelihood (ML) phylogenetic tree of T. nordenskioeldii based on concatenated amino acid sequences of 31 genes from 36 diatom mtDNAs, with Phytophthora ramorum (EU427470) and Saprolegnia ferax (NC_005984) serving as outgroup taxa. The numbers beside branch nodes are the percentages based on 1000 bootstrap replicates.
Acknowledgements
We are thankful to all staffs of marine ecological environment genomics research group in Institute of Oceanology, Chinese Academy of Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW387419, under the accession no. MW387419. The associated BioProject, SRA and Bio-Sample numbers are PRJNA684688 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA684688), SRR13279234 (https://www.ncbi.nlm.nih.gov/sra/SRR13279234) and SAMN17125947 (https://www.ncbi.nlm.nih.gov/biosample/SAMN17125947/), respectively.
Additional information
Funding
References
- Alverson AJ, Beszteri B, Julius ML, Theriot EC. 2011. The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella. BMC Evol Biol. 11:125–125.
- Alverson AJ, Jansen RK, Theriot EC. 2007. Bridging the Rubicon: phylogenetic analysis reveals repeated colonizations of marine and fresh waters by thalassiosiroid diatoms. Mol Phylogenet Evol. 45(1):193–210.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Cleve PT. 1873. On diatoms from the Arctic Sea. Bihang till Kongliga Svenska Vetenskaps-Akademiens Handlingar. 1:1–28.
- Ehara M, Inagaki Y, Watanabe KI, Ohama T. 2000a. Phylogenetic analysis of diatom coxI genes and implications of a fluctuating GC content on mitochondrial genetic code evolution. Curr Genet. 37(1):29–33.
- Ehara M, Watanabe KI, Ohama T. 2000b. Distribution of cognates of group II introns detected in mitochondrial cox1 genes of a diatom and a haptophyte. Gene. 256(1–2):157–167.
- Hoppenrath M, Beszteri B, Drebes G, Halliger H, Van Beusekom JEE, Janisch S, Wiltshire KH. 2007. Thalassiosiraspecies (Bacillariophyceae, Thalassiosirales) in the North Sea at Helgoland (German Bight) and Sylt (North Frisian Wadden Sea) – a first approach to assessing diversity. Eur J Phycol. 42(3):271–288.
- Kaczmarska I, Beaton M, Benoit AC, Medlin LK. 2006. Molecular phylogeny of selected members of the order Thalassiosirales (Bacillariophyta) and evolution of the Fultoportula. J Phycol. 42(1):121–138.
- Kajitani R, Yoshimura D, Okuno M, Minakuchi Y, Kagoshima H, Fujiyama A, Kubokawa K, Kohara Y, Toyoda A, Itoh T. 2019. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat Commun. 10(1):1702.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26(5):589–595.
- Li Y, Zhao Q, Lü S. 2013. The genus Thalassiosira off the Guangdong coast, South China Sea. Botanica Marina. 56(1):83–110.
- Smith SA, Dunn CW. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716.
- Stoermer EF, Julius ML. 2003. Centric Diatoms. In: Freshwater Algae of North America. (Wehr, J.D. & Sheath, R.G. Eds), pp. 559-594. San Diego: Academic Press.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–235.
