Abstract
The complete mitochondrial genome (mitogenome) of Stenopsyche tienmushanensis Hwang, 1957 was assembled from Illumina high-throughput sequencing data. The circularized mitogenome has a length of 16,049 bp with a high A + T content of 77.9%, including 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. Its gene composition and order were the same as Stenopsyche angustata. The overall base composition is 42.0% for A, 35.9% for T, 7.6% for C, and 14.5% for G. A phylogenetic analysis based on 13 PCGs of S. tienmushanensis and 20 other caddisflies supports the placement of Stenopsychidae as sister to the Hydropsychidae within Trichoptera.
Trichoptera, also called caddisflies, is the sixth largest order of Insecta. Caddisflies are widely distributed in the world, and play an important role in water monitoring and evaluation of the health of aquatic ecosystems as bio-indicators (Resh and Unzicker Citation1975). Stenopsychidae is one of the smallest families in Trichoptera, which only has three genera in the world (Morse et al. Citation2014). Stenopsyche is the largest genus of Stenopsychidae, most of which live in clean rivers (Morse et al. Citation2014). The body of Stenopsyche tienmushanensis Hwang, 1957 is yellowish brown, with distinct dark punctate pattern of highly sclerotized head and pronotum (Xu et al. Citation2015). This species widely distributed in China, such as Hubei, Shanxi, Shaanxi, Hunan, Anhui, Zhejiang, Jiangxi, Guizhou and Guangxi Provinces, as well as Hainan Island (Xu et al. Citation2015). Previously studies focused on ecological surveys and the identification of larvae in Stenopsyche genus from China (Tian Citation1985, Citation1988; Tian and Zheng Citation1989). Little is known about the mitogenomes of this genus. To date, whole mitogenome data is available for only one species, Stenopsyche angustata (MT677866.1), in Stenopsychidae (Trichoptera) (Huang et al. Citation2020). Here, we sequence and annotate the complete mitogenome of S. tienmushanensis to facilitate better understanding of the mitochondrial characteristics and the evolutionary history of Stenopsychidae.
In this study, the specimen of S. tienmushanensis was collected from Nanjing, Jiangsu Province, China (32.042°N, 118.848°E). A specimen was preserved at −20 °C and deposited at Nanjing Agricultural University, China (http://www.njau.edu.cn/, Yi Wu, [email protected]) under the voucher number 33 m-8. Genomic DNA was extracted from one leg of a single individual using the QIAamp DNA Micro Kit (Qiagen, GmbH, Germany). The sample was sequenced on the Illumina HiSeq X 10 platform at Novogene (Tianjin, China) following a PE150 strategy. The sequenced mitogenome was assembled by NOVOPlasty v3.8.2 (Dierckxsens et al. Citation2017) and annotated by MitoZ v2.4-alpha (Meng et al. Citation2019).
The circularized mitogenome of S. tienmushanensis has a length of 16,049 bp with a high A + T content of 77.9% (42.0% for A, 35.9% for T, 7.6% for C, and 14.5% for G), under GenBank accession number MW201980. It contained an entire set of 37 genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes) plus a control region. Its gene composition and order were same as S. angustata (Huang et al. Citation2020). All 13 PCGs were AT-biased (75.7% on average), with the highest A + T content (81.4%) in ND6 and the lowest (67.9%) in COX3. The l-rRNA and s-rRNA was 1,380 bp and 876 bp in length, with an A + T content of 83.3% and 85.3%, respectively. All tRNAs were ranging from 65 bp (tRNAHis) to 76 bp (tRNALeu-UUR) in length with typical cloverleaf secondary structure, except for tRNASer-AGN, which has lost the dihydroxyuridine arm. 23 genes (9 PCGs and 14 tRNAs) encoded by the positive strand, and 14 genes (four PCGs, eight tRNAs, and two rRNAs) encoded by the reverse strand. All PCGs were initiated by ATN codons, except for ND4L (GTG). The stop codons of most PCGs were complete (TAA or TAG), except for COX2 and ND5 whose stop codons were incomplete (T).
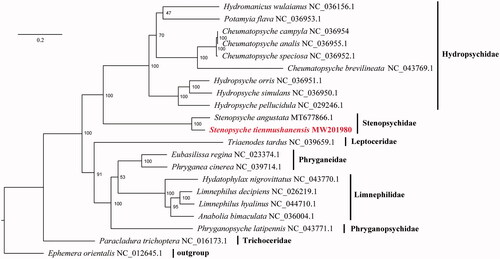
Nucleotide sequences of 13 PCGs from 21 caddisflies were aligned using MAFFT v7.407 (Katoh and Standley Citation2013), and trimmed using trimAl v1.4.1 (Capella-Gutiérrez et al. Citation2009) with heuristic method ‘automated1’. The phylogenetic tree was reconstructed using IQ-TREE v2.0 (Minh et al. Citation2020). The phylogenetic analyses placed S. tienmushanensis as sister to S. angustata with high support (). It supported the results that the monophyly of Stenopsychidae. This monophylum, as well as its placement as sister to Hydropsychidae, which were consistent with the results from previous studies (Huang et al. Citation2020).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW201980. The associated Bio-Sample numbers is SAMN16619568.
References
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Huang J, Wang X, Zhong X, Li Y, Qin H, Wang Y, Wang H. 2020. Characterization of the complete mitochondrial genome of Stenopsyche angustata (Trichoptera, Stenopsychidae). Mitochondr DNA B Resour. 5(3):3114–3115.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Morse JC, Yang L, TianL X. 2014. Trichoptera world checklist. Available from: http://entweb.clemson.edu/database/trichopt/index.htm.
- Resh VH, Unzicker JD. 1975. Water quality monitoring and aquatic organisms: the importance of species identification. J Water Pollut Control Fed. 47(1):9–19.
- Tian LX. 1985. Two new species of genus Stenopsyche McLachlan from Xizang Plateau. J Nanjing Agric Univ. 30(1):23–25.
- Tian LX. 1988. A review of the Chinese genus Stenopsyche McLachlan (Trichoptera: Stenopsychidae). Acta Entomol Sin. 31(2):194–202.
- Tian LX, Zheng J. 1989. Description of two new species of the genus Stenopsyche McLachlan from China (Trichoptera). J Nanjing Agric Univ. 34(12):50–52.
- Xu JH, Sun CH, Wang BX. 2015. A new species of Stenopsyche, with descriptions of larvae and females of some species associated by gene sequences (Insecta: Trichoptera). Zootaxa. 4057(1):63–78.