Abstract
Brassica napus variety NY18 and 088018 are female and male parents of the national registered variety Ningza 1818, respectively. Here, we determined the complete mitochondrial genomes of these two varieties. The genome sizes of NY18 and 088018 were 221,864 bp and 222,015 bp, respectively. Both genomes contained 40 protein-coding genes, 21 tRNA genes, and three rRNA genes. Considerable structural variations existed between the two mitochondrial genomes, which were separated into five syntenic regions. Phylogenetic analysis using the maximum-likelihood method showed that the mitochondrial sequences of B. napus were closely clustered, forming a single clade which had a relatively close relationship with the clade formed by B. rapa, B. juncea, and B. oleracea.
Oilseed rape (Brassica napus Linnaeus 1753. AACC, 2n = 38) is not only the second most important oilseed crop worldwide for human nutrition and industrial products, but also an emerging biofuel crop (Weiss Citation2000). In China, the cultivated area of oilseed rape reaches 67 million hectares, with an annual yield of around 4.5 million tons (Fan et al. Citation2018). The genetic diversity of oilseed rape underlying its massive phenotypic variations remains largely unexplored. But the mitochondrial genome-based phylogenetic analysis would improve our understanding of the evolutionary relationship of this plant and accelerate the genetic improvement of B. napus. Brassica napus variety NY18, selected from variety NY10, is a female parent of the national registered variety Ningza 1818. And 088018 selected from variety NY7 × Marnoo, is a male parent of the national registered variety Ningza 1818 (Fu et al. Citation2015). The characterization and comparison of the complete mitochondrial genomes of varieties NY18 and 088018, and their phylogenetic relationship within Brassicaceae were described in the present study.
Fresh leaves were sampled from the Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu Province (32°2′15″N, 118°52′7″E), China. The voucher specimens were deposited at the herbarium of Key Laboratory of Cotton and Rapeseed, Ministry of Agriculture and Rural Affairs (Feng Chen and [email protected]) under the accession number of NJ2016Y006 and NJ2016Y003 for NY18 and 088018, respectively. Genomic DNAs were extracted using the modified CTAB method. Paired-end libraries of 450 bp were constructed and sequenced on the NovaSeq system (Illumina, San Diego, CA). SMRTbell DNA libraries (∼10 kb) were prepared using the BluePippin size selection system following the official protocol and sent to Shanghai Biozeron Biotech Co., Ltd for Sequel sequencing (PacBio, Menlo Park, CA). More than 5 Gb Illumina data and 300 Mb PacBio data were obtained for each sample (Table S1). The mitochondrial genomes were hybrid assembled using SPAdes 3.13.0 (Antipov et al. Citation2016). Additionally, NOVOplasty 4.0 (Dierckxsens et al. Citation2017) and MITObim 1.9.1 (Hahn et al. Citation2013) were also applied to fill gaps and verify the assembly. Thereafter, the genomes were annotated using GeSeq (Tillich et al. Citation2017), coupled with manual correction.
The complete mitochondrial genomes of B. napus varieties NY18 and 088018 were double-stranded circular molecules with a length of 221,864 bp and 222,015 bp, respectively. Both genomes encoded 40 protein-coding genes, 21 tRNA genes, and three rRNA genes (Table S2). Among those genes, ccmFc, cox2, rps3, and rpl2 contained one intron, nad1 and nad5 contained two introns, nad2 and nad4 contained three introns, and nad7 contained four introns. The genes nad1, nad2, and nad5 were trans-spliced. MUMmer 3.9.2 (Kurtz et al. Citation2004) was employed to compare the two genomes. Considerable structural variations were existed between the NY18 and 088018 mitochondrial genomes, which were separated into five syntenic regions (each >20 kb) (Figure S1). Although the positions of the syntenic regions between the two mitochondrial genomes were rearranged, the nucleotide sequences of these syntenic regions were highly conserved (sequence identity > 99.99% for each region).
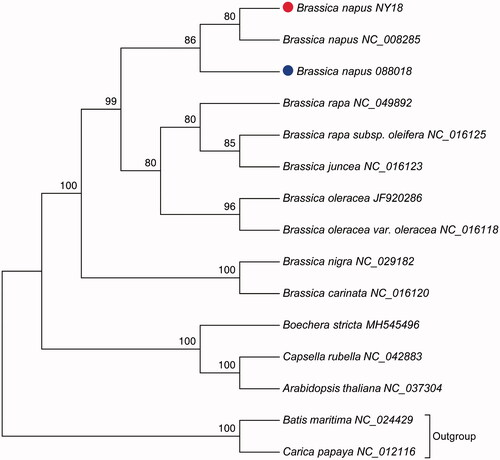
Phylogenetic analysis was performed using 15 complete mitochondrial genomes, and two taxa Batis maritima and Carica papaya were selected as outgroups (Table S3). Whole genome-wide alignments were achieved by HomBlocks pipeline (Bi et al. Citation2018). The maximum likelihood (ML) bootstrap analysis with 1000 replicates was performed using RAxML 8.2.12 (Stamatakis Citation2014). GTR was selected as the best-fit model according to Modeltest3.7 (Posada and Crandall Citation1998). As shown in , phylogenetic tree reveals the topological structure of 15 selected taxa. The three mitochondrial sequences of B. napus were closely clustered, forming a single clade which showed a relatively close relationship with the clade formed by B. rapa, B. juncea, and B. oleracea. The data in this study will provide useful information for further evolutionary analysis of Brassica.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW001149 for NY18 and MW348924 for 088018. The associated BioProject accessions of NY18 and 088018 are PRJNA662838 and PRJNA682878, respectively.
Additional information
Funding
References
- Antipov D, Korobeynikov A, McLean JS, Pevzner PA. 2016. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 32(7):1009–1015.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fan C, Tian J, Hu Z, Wang Y, Lv H, Ge Y, Wei X, Deng X, Zhang L, Yang W. 2018. Advances of oilseed rape breeding (in Chinese with an English abstract). J Plant Genet Resour. 19(3):447–454.
- Fu SX, Qi CK, Gu H, Zhang JF, Pu HM, Chen S, Zhou XY, Chen F, Gao JQ, Long WH, et al. 2015. Breeding of high-quality two-line hybrid rape variety Ningza 1818 through chemical emasculation (in Chinese with an English abstract). Acta Agric Jiangxi. 27(7):11–14.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Weiss EA. 2000. Oilseed crops. London (UK): Blackwell Publishing Limited.