Abstract
In this study, the complete mitochondrial genome of Hippopus porcellanus was reported. The whole mitochondrial genome was 21,565bp in length with a typical mitochondrial genomic structure including 13 protein-coding genes, 23 transfer RNA genes, 2 ribosomal RNA genes and 1 control region (D-loop). Mitogenome base composition was biased toward A + T content, at 60.3%. A phylogenetic tree based on complete mitogenome sequences revealed that, H. porcellanus is closely related to H. hippopus, both of which belong to the genus Hippopus.
Giant clams (Bivalvia: Tridacnidae), the largest bivalve mollusks in the world, inhabit the shallow coral reefs of the Indo-Pacific region. Giant clams are not only important to coral reef ecosystems, but also constitute a significant food source in Asia and the South Pacific, and are in demand for the shell and aquarium trade (Keys and Healy Citation1999). These clams belong to the subfamily Tridacninae, which has two genera namely: Hippopus and Tridacna (Othman et al. Citation2010). Two species belong to Genus Hippopus: Hippopus hippopus Linnaeus 1758 and Hippopus porcellanus Rosewater 1982. In this study, we sequenced the complete mitochondrial genome of H. porcellanus to further investigate of the taxonomy and phylogenetic relationships of Tridacnidae by increasing the amount of available molecular data.
The specimen was collected from Sanya, Hainan province, China (N109.51, E18.21) by a local fisherman, and stored in Tropical Marine Biodiversity Collections of South China Sea (TMBC), Chinese Academy of Sciences, Guangzhou, China (specimen accession number: TMBC030713). The total genomic DNA was extracted following the modified CTAB DNA extraction protocol (Attitalla Citation2011), and followed by library preps and pair-end sequenced (2 × 150 bp) with HiSeq (Illumina, San Diego, CA). Approximately 5,987 Mb of raw data and 5,216 Mb of clean data were obtained, and de novo assembled by the SOAP de novo software (Zhao et al. Citation2011) with an average of approximately 310× coverage.
The mitogenome of H. porcellanus was 21,565bp in length (GenBank accession number MT755622), containing 13 protein-coding genes (PCGs), 23 transfer RNA genes (tRNAs), two ribosomal RNA (12S rRNA and 16S rRNA) genes and a non-coding control region (D-loop). The mitogenome base composition of H. porcellanus was biased toward A + T content at 60.3% (26.4% A, 33.9% T, 15.0% C, 24.6% G). The 13 identified PCGs vary in length from 114 to 1,677 bp. COI, ATP8, ND5, ND2, ND4L, ND1 and ATP6 initiated with ATG as the start codon, while ND3 and COIII begin with TTG, ND4 begin with ATT, COII with ATA, ND6 with GTG and Cytb with ATC. Three types of stop codons were TAA (ND4, COII, COIII, ND4L, ND6, Cytb), TAG (COI, ATP8, ND5, ATP6, ND3, ND1) and TAT (ND2). The lengths of the 23 tRNA genes ranged from 58 to 74 bp, and all of the tRNA genes contained typical secondary structure. The 12S rRNA gene was located between tRNA-Leu and tRNA-Second Leu, and was 922 bp long, while the 16S rRNA gene was located between tRNA-Ile and ND1, with a length of 1,269 bp. A 2,605 bp control region (D-loop) was located between tRNA-Met and COII, with an A + T content of 54.7%.
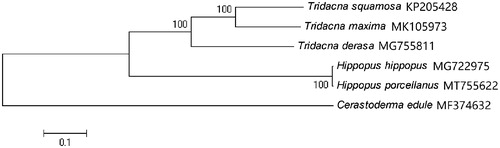
A neighbor-joining phylogenetic tree of H. porcellanus with five other closely related species was constructed with the complete mitochondrial genomes using MEGA6 (Tamura et al. Citation2013) (). The result suggested that, H. porcellanus is closely related to H. hippopus, both of which belong to the genus Hippopus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT755622. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA704757, SRR13827840, and SAMN18054235, respectively.
Additional information
Funding
References
- Attitalla IH. 2011. Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J Biol Sci. 14(21):998–999.
- Keys JL, Healy JM. 1999. Sperm ultrastructure of the giant clam Tridacna maxima (Tridacnidae: Bivalvia: Mollusca) from the Great Barrier Reef. Mar Biol. 135(1):41–46.
- Othman AS, Goh GHS, Todd PA. 2010. The distribution and status of giant clams (Family Tridacnidae) – a short review. The Raffles Bulletin of Zoology. 58(1):103–111.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2272.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.