Abstract
In this study, the complete mitochondrial genome of Schizoneuraphis gallarum was sequenced and annotated. The mitogenome is 14,990 bp in length with an A + T content of 82.6%, including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and a control region. This is the shortest aphid mitogenome reported so far. All the protein-coding genes are initiated with ATN codon and terminated with TAA codon except cox1 and nad4 which are terminated with T. All the transfer RNA genes can form the typical clover-leaf secondary structure except trnS (AGN). The maximum-likelihood phylogenetic tree based on mitogenome sequences of 25 aphid species shows that S. gallarum is sister to Hormaphis betulae.
Keywords:
Aphid species of the subfamily Hormaphidinae are characterized by seasonally migrating between primary and secondary host plants, inducing galls on the primary host plants, secreting wax and producing soldier aphids (Chen et al. Citation2014). To date, five complete mitochondrial genome sequences of three Hormaphidinae species have been reported, none of which are mitogenomes of aphids belonging to the tribe Nipponaphidini. In this study, we sequenced and annotated the complete mitochondrial genome of the first representative of Nipponaphidini, Schizoneuraphis gallarum van der Goot, 1917 (GenBank accession number MT822308). Schizoneuraphis gallarum is mainly distributed in southeastern Asia and migrates between the primary host plant Distylium stellare Kuntze, 1891 where balloon-shaped galls are formed and secondary host plants Litsea Lamarck, 1791 (Blackman and Eastop Citation2020). The samples of S. gallarum were collected on Litsea sp. from Mt. Daming, Guangxi, China (23.5244°N, 108.3444°E) and deposited in the National Zoological Museum of China, Institute of Zoology, Chinese Academy of Sciences, Beijing, China (NZMC no. 26139, Jing Chen, [email protected]).
The mitogenome of S. gallarum was amplified using short and long PCR reactions (Wang et al. Citation2016) and sequenced on an ABI 3730 automated sequencer. The mitogenome is 14,990 bp long, which is the shortest aphid mitogenome published so far. It contains 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a control region. The genes are arranged in the same order as the inferred ancestral insect mitochondrial genome (Clary and Wolstenholme Citation1985). There are 11 gene intervals and 16 overlaps among the mitogenome of S. gallarum. The nucleotide composition of the whole mitogenome is 37.9% T, 11.5% C, 44.7% A, and 5.9% G, and the A + T content is 82.6%.
All the PCGs start with the typical start codon ATN. Eleven PCGs end with TAA codon, while cox1 and nad4 are terminated with a single T residue. The lengths of 22 tRNAs range from 62 to 73 bp. All the tRNAs can form the typical clover-leaf secondary structure except trnS (AGN) of which the dihydrouridine (DHU) arm is lost. The gene rrnL is 1267 bp long and rrnS is 769 bp long. The control region located between rrnS and trnI is 473 bp in length and its A + T content is 83.3%. The control region consists of an AT-rich region, a poly-thymidine stretch and a stem-loop structure region.
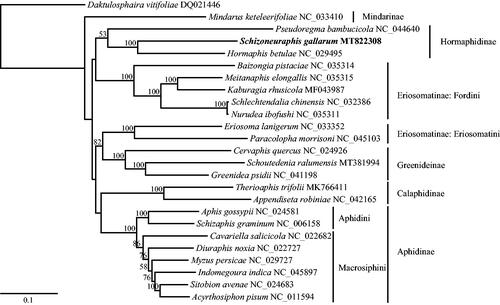
A maximum-likelihood phylogenetic tree was constructed using RAxML v8.2.10 (Stamatakis Citation2014) based on the whole mitogenomes of S. gallarum and 24 other aphid species. All subfamilies including more than one representative species were monophyletic except Eriosomatinae. Within the clade of Hormaphidinae, S. gallarum was positioned as the sister to Hormaphis betulae (Mordvilko, 1901) with strong support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MT822308.
Additional information
Funding
References
- Blackman RL, Eastop VF. 2020. Aphids on the word’s plants. [accessed 2020 Sep 9]. http://www.aphidsonworldsplants.info/.
- Chen J, Jiang LY, Qiao GX. 2014. A total-evidence phylogenetic analysis of Hormaphidinae (Hemiptera: Aphididae), with comments on the evolution of galls. Cladistics. 30(1):26–66.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang Y, Jiang LY, Liu YJ, Chen J, Qiao GX. 2016. General methods to obtain and analyze the complete mitochondrial genome of aphid species: Eriosoma lanigerum (Hemiptera: Aphididae) as an example. Zoological Systematics. 41(2):123–132.