ABSTRACTS
Aquilaria species is one of the main plant resources that produce agarwood, which containing black resin with important economic and medicinal values. There are about 15 species known to the genus around the world, but only two can be found in China, i.e. A. sinensis and A. yunnanensis. In this study, A. sinensis and A. yunnanensis that endemic respectively to Hainan and Yunnan were sampled, on the basis of the investigation and observation of their main morphological features in plantation. Five primers, i.e. ITS2, matK, trnL-trnF1, trnL-trnF2, and trnH-psbA, were eventually selected for DNA barcoding. The results showed that the seed surface of A. sinensis is smooth or sparsely pubescent, and the seed appendages were long. While the seed surface of A. yunnanensis is densely covered with yellow hairs and the seed appendages are short. The trnL-trnF1 sequence fragment has significant intraspecific and interspecific genetic distances. However, the species identification success rate of ITS2+matK combination was finally screened to be the highest, which was verified by the BBA method of TaxonDNA. The phylogenetic trees cluster analysis revealed that the classification of A. sinensis and A. yunnanensis is significant, and there is geographic isolation between the two species. Therefore, on the premise of accurate identification of plant morphological characters, ITS2+matK combination can be used to accurately identify the Aquilaria species in China.
Introduction
Aquilaria species of Thymelaeaceae, the tropical and subtropical evergreen trees, that are mainly distributed in tropical or subtropical regions of the Southeast Asia, are the most important plant resources for the production of rare agarwood (FOC Eco Citation1999). Agarwood is a precious traditional medicinal ingredient and natural perfume in China, and it has been used widely for cultural, religious, and medicinal purposes around the world. In recent years, wild resources of Aquilaria species are found depleted due to the serious human logging and the destruction of their natural environments. Aquilaria crassna is listed as a critically endangered species by International Union for Conservation of Nature (IUCN), and A. malaccensis, as well as A. sinensis, are also regarded as vulnerable species (Hashim et al. Citation2016). In addition, all Aquilaria species have been included in the Convention on International Trade in Endangered Species (CITES) ((CITES) Citation2004).
Because of the high level of similarity in morphological features of Aquilaria species, previous identification methods are only dependent on the classifications of the different morphological features of flower, seed, and fruit. In addition, the flowering and fruiting period of Aquilaria species is unstable, and its wild resources are on the verge of extinction. Therefore, it is extremely difficult to identify the Aquilaria species by only collecting their fruits and flowers through field sampling, not to mention the high error generated (Lee and Mohamed Citation2016). In conclusion, this is also one of the factors that have not been clear about the taxonomic study of Aquilaria species. Several studies have shown that more than 20 Aquilaria species are distributed in the tropical regions of Southeast Asia (Lee et al. Citation2016; Gao et al. Citation2017). The Flora of Malaysia is one of the earliest Flora to describe the morphological characteristics of Aquilaria species (Hou Citation1960). And Aquilaria species are divided into 12 taxons in this flora, i.e. A. malaccensis, A. microcarpa, A. brachyantha, A. urdanetensis, A. citrinaecarpa, A. apiculata, A. filaria, A. parvifolia, A. hirta, A. rostrata, A. beccariana, and A. cumingiana. Over the past few years, Aquilaria species in Asian mainland were indicated that can be divided into 13 taxons (Santisuk Citation2007), i.e. A. baillonii, A. banaensis, A. beccariana, A. crassna, A. hirta, A. khasiana, A. malaccensis, A. microcarpa, A. rostrata, A. rugosa, A. sinensis, A. subintegra, and A. yunnanensis. However, about 15 Aquilaria species that discovered across the world were recorded in Flora of China (FOC Eco Citation1999), including A. sinensis and A. yunnanensis.
DNA barcoding has been proved the quick and accurate approach to identify different species based on selection of standard DNA segments (Hebert et al. Citation2003). This technology are also used widely for the identification of Aquilaria species. For example, trnL-trnF sequence was found that can provide new molecular framework for the identification of Aquilaria species (Eurlings and Gravendeel Citation2005). Similarly, phylogenetic trees constructed by trnL-trnF+ITS2 combination was demonstrated that is useful for identifying the Aquilaria species (Lee et al. Citation2016). Meanwhile, ITS sequence was applied in first time to analyze A. malaccensis from different sources (Lee et al. Citation2018). In addition, phylogenetic tree constructed by trnL-trnF+ITS1 was found that could aggregate the DNA sequence of A. sinensis in GenBank (Jiao et al. Citation2014). When comparing A. sinensis, A. yunnanensis and A. crassna, Li et al. found that the combinations of ITS+matK and ITS+trnL-trnF are suitable for molecular identification of these three species (Li et al. Citation2018). In the previous research, our group also found that matK fragments play an important role in Aquilaria species from multiple sources (Kang et al. Citation2019).
Although DNA barcoding is crucial in the identifications of various Aquilaria species, the selection of barcode fragments or combinations may be different for different materials. Moreover, DNA barcode technology combined with traditional classification features can obtain the best identification results on the basis of accurate collection of samples. Hence, 18 samples of A. sinensis and seven of A. yunnanensis were collected respectively from six plantations in Hainan and two plantations in Yunnan. Both Aquilaria species were studied on site during their flowering and fruit-bearing periods, and their morphological features were summarized. Five primers were selected for DNA barcodes study. On the premise of determining the morphological characteristics, it is planned to screen out the barcode fragments or combinations suitable for the identification and the construction of phylogenetic trees of both Aquilaria species by analyzing the sequence characteristics, genetic distances and species identification rates of different primers.
Materials and methods
Materials
A total of 25 samples of A. sinensis and A. yunnanensis that collected from 9 plantations in Hainan and Yunnan of China were used as the experimental materials. Fresh leaves were dried and preserved with silica gel, then extracted the total plant DNA. The voucher specimens were also deposited in the Herbarium of Traditional Chinese Medicine, Hainan Branch of the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences. Localities of all sampled accessions are shown in .
Table 1. Sample collection information and GenBank accession numbers of the Aquilaria species generated through this study.
Methods
Observation of the main morphological features of Aquilaria species
The six plantations in Hainan and two plantations in Yunnan were investigated. Description of the main reproductive organ characteristics, such as fruits and seeds of A. sinensis and A. yunnanensis by collecting samples of the Aquilaria species. And it was made into wax leaf specimens for preservation.
DNA extraction, amplification and sequencing
The total DNA extraction kit for plants that acquired from Tiangen Biotech (Beijing) Co., Ltd was used to extract the DNA, and five primers (i.e. ITS2, matK, trnL-trnF1, trnL-trnF2 and trnH-psbA) were used for PCR amplification (). Optimization and adjustment were made based on the previously reported PCR reaction system (Group et al. Citation2009). The sequencing of all amplification products was completed by Guangzhou IGE Biotechnology Ltd. Bioedit (Hall Citation1999), Sequencematrix (Vaidya et al. Citation2011), Mega X (Sudhir et al. Citation2018), MrBayes 3.2.6 (Huelsenbeck and Ronquist Citation2001) and PAUP 4 b (http://paup.phylosolutions.com) were used to edit and compare the sequences, match the barcodes, calculate the genetic distances and build the phylogenetic trees, while TaxonDNA (Meier et al. Citation2006) was used to calculate species identification rates, and R 4.0.0 (https://www.r-project.org) and Figtree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) were used to beautify the phylogenetic trees.
Table 2. Details on the PCR primers used in this study.
Data analysis
The rate of PCR amplification can be defined as the ratio in the percentage of the number of successful individuals to the total number of individuals while sequencing success rate is the percentage of the number of high-quality sequences obtained to the total number of sequences (Kress et al. Citation2009). MEGA X was used to compare multiple sequences, calculate sequence length, variable sites and conserved sites. Intraspecific and interspecific genetic distances were calculated by K2P model of MEGA X. The ‘Best match,’ ‘Best close match’ and ‘All species barcodes’ (BBA method) in TaxonDNA software were used to evaluate the success rates of species identification and to screen for the best DNA fragments or combinations. Moreover, the phylogenetic trees were generated using the neighbor-joining (NJ) and Unweighted Pair-group Method with Arithmetic Mean (UPGMA) methods in MEGA X, with individual node support calculated based on 1000 bootstrap re-samplings. In addition, Bayesian interference (BI) and Maximum Likelihood (ML) approaches were also chosen for the construction of phylogenetic trees in MrBayes 3.2.6 and PAUP 4 b, respectively.
Results
Main morphological features of A. sinensis and A. yunnanensis
Fruit of A. sinensis is oblong, with a long beak, moderate calyx without wrapping the fruit, and its seed surface is smooth or sparsely covered with white pubescence, with long appendages. However, fruit of A. yunnanensis is oval, with small and scattered calyx, short seed appendages, and the seed surface is densely covered with yellow pubescence. In conclusion, the main distinguishing characteristics between A. sinensis and A. yunnanensis are whether the seed surface is densely covered with yellow pubescence and the length of seed appendages ( and ).
Distinguishing both Aquilaria species using DNA barcoding
Sequence characteristics of the DNA barcodes
PCR amplification and sequencing were implemented for ITS2, matK, trnL-trnF1, trnL-trnF2 and trnH-psbA sequences of all samples to obtain the corresponding rates of PCR amplification, sequencing rates, and variable sites data (). It could be observed from the Table3 that the rates of PCR amplification and sequencing rates of above 5 sequences were 100.00%. Ordering their lengths in a descending order, matK (775) > ITS2 (456) = trnL-trnF1 (456) > trnH-psbA (408)> trnL-trnF2 (115).The number of variant sites from highest to lowest is trnL-trnF1 (11) > ITS2(2) > matK (1) > trnL-trnF2 (0) = trnH-psbA (0), while the number of conserved sites from highest to lowest is matK (769) > ITS2 (450) > trnL-trnF1 (439) > trnH-psbA (407) > trnL-trnF2 (115).The number of singleton sites in trnL-trnF1 was 4, and the rest of the fragments were 0.
Table 3. Evaluation of the five DNA barcode loci and their combinations.
Genetic distance
The K2P distance model was selected by MEGA X software to calculate the interspecific and intraspecific genetic distances among five fragments and their combinations. The results are as shown in Supplementary Table 1. For A. sinensis, the range of intraspecific genetic distances among the five primers and their combinations was 0.000–0.016%, while the range of average intraspecific genetic distances was 0.000–0.007%. For A. yunnanensis, the range of intraspecific genetic distances among the five primers and their combinations was 0.000–0.016%, while the range of average intraspecific genetic distances was 0.000–0.006%. Moreover, the range of interspecific genetic distances between two species was 0.000–0.016%, while the range of average intraspecific genetic distances was 0.000–0.006%. To summarize, this study found that trnL-trnF1 has the maximum average intraspecific and interspecific genetic distances, followed by ITS2.
Species identification
This paper used the ‘BBA’ method in TaxonDNA to verify and analyze the identification rates for the Aquilaria species, and the results are as shown in . The best performing single fragment in ‘best match’ and ‘best close match’ was matK, with an accurate identification rate of 81.00%. Some of the best performing multi- fragment combinations in ‘best match,’ ‘best close match,’ and ‘all species’ include ITS2+matK (96.00%), matK+trnL-trnF2 (81.00%), matK+trnH-psbA (81.00%), ITS2+matK+trnL-trnF1 (84.00%), ITS2+matK+trnL-trnF2 (96.00%), ITS2+matK+trnH-psbA (96.00%), matK+trnL-trnF2+trnH-psbA (81.00%), ITS2+matK+trnL-trnF1+trnL-trnF2 (84.00%), ITS2+matK+trnL-trnF1+trnH-psbA (84.00%), ITS2+matK+trnL-trnF2+trnH-psbA (96.00%), and trnL-trnF1+trnL-trnF2+trnH-psbA (84.00%).
Phylogenetic tree
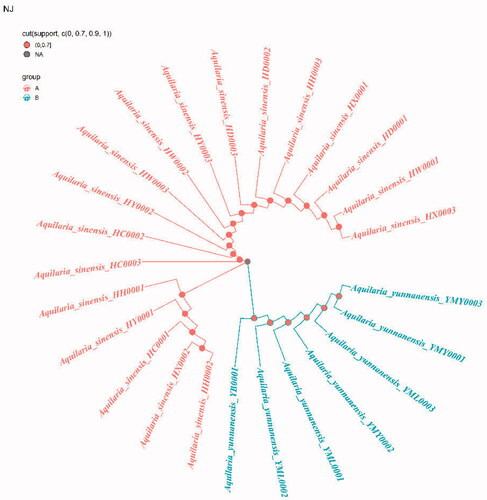
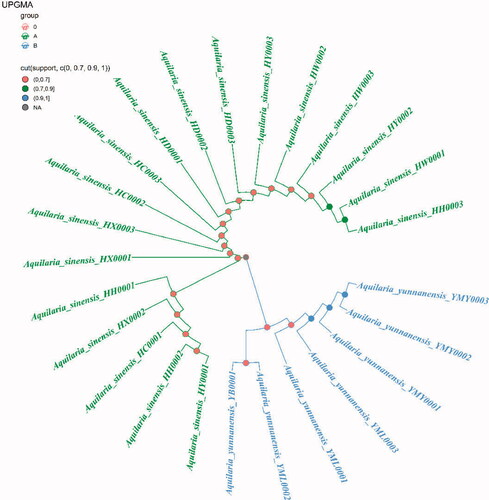
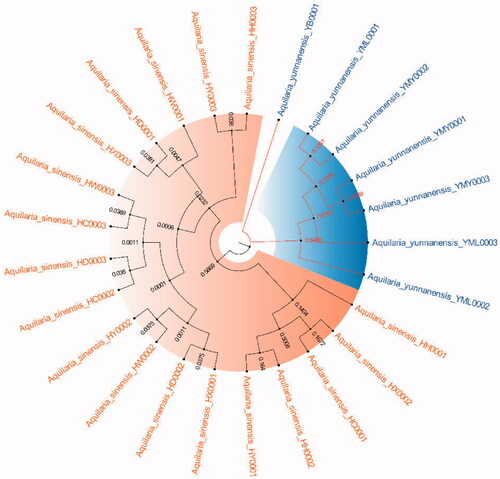
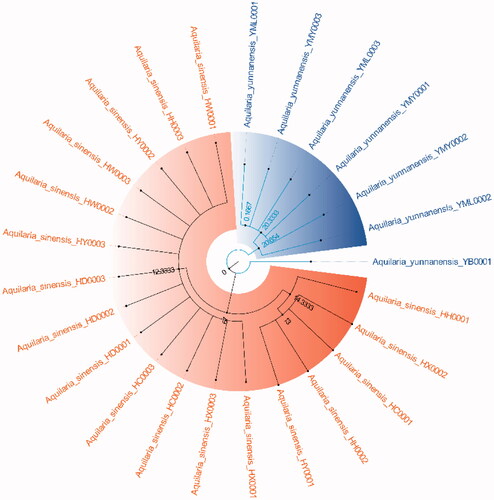
Because ITS2+matK, ITS2+matK+trnl-trnf2, ITS2+matK+trnH-psbA and ITS2+matK+trnl-trnf2+trnH-psbA have the highest success rates of species identification (96%) and the 2 fragment combinations are more convenient in the process of constructing phylogenetic tree, with the sequencing cost was low, so this study chose ITS2+matK combination to construct NJ, UPGMA, BI and ML phylogenetic trees respectively (). The results showed that the phylogenetic trees constructed by the four methods can clearly divide A. sinensis from Hainan and A. yunnanensis from Yunnan, with significant geographic isolation between the two species.
Discussion
DNA barcoding evaluation of Aquilaria species in China
This study used five primers (i.e. ITS2, matK, trnL-trnF1, trnL-trnF2 and trnH-psbA) to analyze the DNA barcodes of A. sinensis and A. yunnanensis (). In 2009, rbcL and matK were proposed officially as universal barcodes for terrestrial plants by CBOL research team. Then, ITS and trnH-psbA, which demonstrate faster evolutionary rates, were suggested as candidate barcodes by all participants in the 3rd International Academic Conference of DNA Barcode (Group et al. Citation2009). That is, ITS, matK, rbcL and trnH-psbA are considered as common DNA barcodes. However, the coding sequence of rbcL is highly conserved, leading its variation mainly exists at the level of genus or above and is usually small at the species level (Newmaster et al. Citation2006; Kress and Erickson Citation2007; Lahaye et al. Citation2008). By filtering DNA barcodes from medical biology and analyzing 6600 ITS2 sequences of 4800 algae, fungus and higher plants under 753 genera, Chen Shilin’s team found that the resolution success rate of ITS2 at the species level is 92.7%. They proposed that ITS2 could be used as a new DNA barcode for fungi and green plants (Chen et al. Citation2010). Furthermore, ITS2 has a shorter sequence length than ITS, and its high rates of PCR amplification or sequencing (Wang et al. Citation2016). Meanwhile, trnL-trnF was also found that plays an important role in molecular identification for Aquilaria species (Eurlings and Gravendeel Citation2005). Therefore, ITS2, matK, trnH-psbA, trnL-trnF1 and trnL-trnF2 were selected for the identification of Aquilaria species in this study.
By comparing the sequence characteristics of different primers and analyzing their genetic distances and species identification rates, we found that the ITS2+matK barcode combination has the highest species identification success rate (96%), which could be apply to identification and phylogenetic tree construction of A. sinensis and A. yunnanensis (). DNA barcoding has been widely for identifying Aquilaria species in nowadays. For example, Jiao et al. found that the significant clustering effect of phylogenetic tree construction by trnL-trnF and ITS1 for A. sinensis (Jiao et al. Citation2014). When analyzing Aquilaria species, Lee et al. concluded that the phylogenetic tree constructed with combination ITS2+trnL-trnF was applicable for Aquilaria species (Lee et al. Citation2016). Li et al. found that the phylogenetic tree constructed by ITS+matK and ITS+trnL-trnF is beneficial to the identification of three Aquilaria species (Li et al. Citation2018). In this study, our results are differrent from those previous studies due to different fragments or combinations tend to have different identification effects for various Aquilaria species. Although the trnL-trnF1 sequence showed significant intraspecific and interspecific genetic distances, the fragment and its combination displayed low species identification rates (). This may result from some repeated sequences in trnL-trnF1 sequencing results, which is not useful for constructing the phylogenetic tree. Moreover, trnL-trnF2 is not applicable for phylogenetic tree construction in this study due to the short sequence length, insignificant genetic distance, and low species identification rate. By analyzing the Aquilaria species in different countries, our the research team found that the matK fragment plays an important role in identifying Aquilaria species (Kang et al. Citation2019). And matK (81%) also showed the highest species identification rate in this study. However, the combination of two fragments is more convenient to operate and reduce the cost of sequencing, and the trnH-psbA variable sites is low. (Kress et al. Citation2005). For these reasons, ITS2+matK was finally selected from the four combinations (i.e. ITS2+matK (96.00%), ITS2+matK+trnL-trnF2 (96.00%), ITS2+matK+trnH-psbA (96.00%), ITS2+matK+trnL-trnF2+trnH-psbA (96.00%)) and used for the clustering analysis of phylogenetic tree for the Aquilaria species in this study.
Morphological difference and study progress of A. sinensis and A. yunnanensis
This study found that A. sinensis has either smooth surface or is grown with sparse hair and long appendages on its seeds through field investigation, while A. yunnanensis has dense yellow pubescence and short appendages on its seeds, which are consistent with the main identification features as recorded in Flora of China. For example, the texture of the capsule of A. sinensis is slightly thin, the skin does not shrink when it is dry, the seeds are white silky or glabrous, the apex has a long beak, the base appendage is longer, about 1.5 cm, longer than the seed. However, A. yunnanensis has thick capsules, shriveled dry pericarps, yellow pubescence on the seeds, short floral organs on top, and short base appendages that are almost equivalent with that of the seeds, with a length of about 0.8–1 cm (FOC Eco Citation1999). In addition, A. sinensis is mainly distributed in Guangdong (including HK and Macao), Hainan and Guangxi, while A. yunnanensis is mainly distributed in Xishuangbanna, Yunnan (Huang Citation1985).
The phylogenetic tree that constructed by ITS2+matK combination has clearly branched out A. sinensis and A. yunnanensis (), showing their distant genetic relationship, which is speculated to have resulted from geographical isolation. At present, the research of A. sinensis has involved molecular, genetic, microscopic, chemical and aromatic aspects (Liu Y et al. Citation2013; Liu P et al. Citation2019; Sun et al. Citation2020; Wang J et al. Citation2019; Wang Z et al. Citation2020). A. sinensis is a unique source of domestic agarwood according to Chinese Pharmacopeia (Committee SP Citation2020). However, there are relatively scarce studies that focus on A. yunnanensis. Although they are greatly different in terms of the morphological features of seeds or fruits, the anatomical structures of A. yunnanensis are basically consistent with A. sinensis, with endophloem scattered in its xylem (Su et al. Citation2016). This enables A. yunnanensis to generate agarwood for making agarwood for medicine or spice. However, whether there are differences in the regulatory genes or chemical components of the two Aquilaria species still needs further study in the process of forming agarwood. And there is a new record of A. yunnanensis in Vietnam (Van Sam et al. Citation2019).
Most Aquilaria species are distributed mainly in tropical or subtropical regions of Southeast Asia, leading in great challenges for taxonomical study. However, DNA barcode technology can ensure the reliability of the results when the sampling is accurate and covers a wide range of distribution (Ren and Chen Citation2010). Therefore, traditional taxonomy is essential for accurately collecting and identifying the Aquilaria species. Moreover, the investigation and collection of Aquilaria species is the key to identify the original species of agarwood, and this work can promote the stability of agarwood market.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov), and the reference numbers [MW118060-MW118109, MW124309-MW124383] are shown in .
Additional information
Funding
References
- (CITES) CoITiESoWFaF. 2004. Consideration of proposals for amendment of appendices I and II – Aquilaria spp. and Gyrinops spp. Thirteenth meeting of the Conference of the Parties, Bangkok, Thailand, 2–14 October.
- Chen S, Yao H, Han J, Liu C, Song J, Shi L, Zhu Y, Ma X, Gao T, Pang X, et al. 2010. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 5(1):e8613.
- Committee SP. 2020. Pharmacopoeia of the people's Republic of China. Beijing: China Pharmaceutical Science and Technology Press.
- Eurlings M, Gravendeel B. 2005. TrnL-trnF sequence data imply paraphyly of Aquilaria and Gyrinops (Thymelaeaceae) and provide new perspectives for agarwood identification. Plant Syst Evol. 254(1–2):1–12.
- FOC Eco. 1999. Flora of China. Vol. 52. Beijing: Science Press.
- Gao Z, Zhao W, Sun P, Wei J. 2017. Species and conservation status of the endangered agarwood-producing genus Aquilaria. Mod Chin Med. 19(08):1057–1063.
- Group CPW, Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL. 2009. A DNA barcode for land plants. Proc Natl Acad Sci USA. 106(31):12794–12797.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41(41):95–98.
- Hashim YZ, Kerr PG, Abbas P, Mohd Salleh H. 2016. Aquilaria spp. (agarwood) as source of health beneficial compounds: A review of traditional use, phytochemistry and pharmacology. J Ethnopharmacol. 189:331–360.
- Hebert PD, Cywinska A, Ball SL, Dewaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond B. 270(1512):313–321.
- Hou D. 1960. Thymelaeaceae. Flora Malesiana-Series 1. Spermatophyta. 6(1):1–48.
- Huang S. 1985. Taxa nova Thymelaeacearum sinicarum. Acta Botanica Yunnanica. 03:277–291.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Jiao L, Yin Y, Cheng Y, Jiang X. 2014. DNA barcoding for identification of the endangered species Aquilaria sinensis: comparison of data from heated or aged wood samples. Holzforschung. 68(4):487–494.
- Kang Y, Liu Y, Yang Y, Feng J, Zheng X, Wei J. 2019. Screening of DNA barcoding sequences for identification of multiple species of Aquilaria L. Chin Pharm J. 54(23):1926–1932.
- Kress WJ, Erickson DL. 2007. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2(6):e508.
- Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E. 2009. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc Natl Acad Sci U S A. 106(44):18621–18626.
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. 2005. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA. 102(23):8369–8374.
- Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V. 2008. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA. 105(8):2923–2928.
- Lee S, Mohamed R. 2016. Rediscovery of Aquilaria rostrata (Thymelaeaceae), a species thought to be extinct, and notes on Aquilaria conservation in Peninsular Malaysia. Blum - J Plant Tax and Plant Geog. 61(1):13–19.
- Lee SY, Mohamed R, Faridah-Hanum I, Lamasudin DU. 2018. Utilization of the internal transcribed spacer (ITS) DNA sequence to trace the geographical sources of Aquilaria malaccensis Lam. populations. Plant Genet Resour. 16(2):103–111.
- Lee SY, Ng WL, Mahat MN, Nazre M, Mohamed R. 2016. DNA barcoding of the endangered Aquilaria (Thymelaeaceae) and Its application in species authentication of agarwood products traded in the market. PLoS One. 11(4):e0154631.
- Li Q, Yan H, Lin D, Wang Y, He M, Zhang W, Gao X, Zhu S. 2018. Molecular identification of three Aquilaria (Thymelaeaceae) species through DNA barcoding. Biol Pharm Bull. 41(6):967–971.
- Liu Y, Chen H, Yang Y, Zhang Z, Wei J, Meng H, Chen W, Feng J, Gan B, Chen X, et al. 2013. Whole-tree agarwood-inducing technique: an efficient novel technique for producing high-quality agarwood in cultivated Aquilaria sinensis trees. Molecules. 18(3):3086–3106.
- Liu P, Zhang X, Yang Y, Sui C, Xu Y, Wei J. 2019. Interxylary phloem and xylem rays are the structural foundation of agarwood resin formation in the stems of Aquilaria sinensis. Trees Struct Funct. 33(2):533–542.
- Meier R, Shiyang K, Vaidya G, Ng PKL. 2006. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol. 55(5):715–728.
- Newmaster SG, Fazekas AJ, Ragupathy S. 2006. DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Botany. 84(3):335–341.
- Ren B, Chen Z. 2010. Plant DNA barcode technology. Bull Bot. 45(01):1–12.
- Van Sam H, The Nha N, Van Chu T, Thanh Tuan N, Thi Tho N, Thanh Tam D, Bao Thanh L, Ngoc Hai T, Van Huan H, Thi Hang H, et al. 2019. Aquilaria yunnanensis S.C. Huang (Thymelaeaceae), a new record for the flora of Vietnam. FS. 3(2):202–208.
- Sang T, Crawford DJ, Stuessy TF. 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot. 84(8):1120–1136.
- Santisuk T. 2007. Taxonomy geography and ecology of Aquilaria Lamk (Thymelaeaceae) in continental Asia. Second International Agarwood Conference March 5–6, Bangkok, Thailand.
- Su J, Liu Z, Li R, Ren C, Yang X. 2016. Preliminary study on timber structure of Aquilaria yunnanensis. Chin J Trop Agric. 36(04):30–34.
- Sudhir K, Glen S, Michael L, Christina K, Koichiro T. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Sun P-W, Xu Y-H, Yu C-C, Lv F-F, Tang X-L, Gao Z-H, Zhang Z, Wang H, Liu Y, Wei J-H. 2020. WRKY44 represses expression of the wound-induced sesquiterpene biosynthetic gene ASS1 in Aquilaria sinensis. J Exp Bot. 71(3):1128–1138.
- Tate JA, Simpson BB. 2003. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot. 28(4):723–737.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Wang Z, Cao H, Cai C, Wang Z. 2020. Using genetic markers to identify the origin of illegally traded agarwood-producing Aquilaria sinensis trees. Global Ecol Conserv. 22:e00958.
- Wang J, Wang Y, Yan J, Li W, Dong W, Mei W, Dai H. 2019. Comparison of the anatomy structure and chemical compositions of agarwoods from two kinds of Aquilaria sinensis. Sci Silva Sin. 55(07):146–154.
- Wang XY, Zheng SH, Liu Y, Han JP. 2016. ITS2, a better DNA barcode than ITS in identification of species in Artemisia L. Chinese Herbal Medicines. 8(4):352–358.