Abstract
The complete mitochondrial genome of the fine flounder Paralichthys adspersus, was determined for the first time through Next Generation Sequencing (NGS) approach. The mitogenome (GenBank accession no. MW288827) has 17,060 bp in length and consisted of the well-known 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and the control region. The overall nucleotide composition of the whole mitogenome was A: 27.5%, C: 29.5%, G: 17.1%, and T: 25.9%. Phylogenetic analyses based on 12 protein-coding genes clustered P. adspersus in the monophyletic Paralichthyidae clade, showing the closest phylogenetic relationship with its congeneric species P. olivaceus.
Keywords:
The fine flounder, Paralichthys adspersus (Steindachner, 1867), is a commercially important species, which is distributed on the Pacific coast from Ecuador to Chile (Chirichigno and Cornejo Citation2001). In Peru, this species not only supports the most important artisanal flatfish fishery but also has great potential for aquaculture to local and international markets due to the high demand for its exquisite white flesh. Notwithstanding, the mitochondrial genome of P. adspersus remains unknown and to date, there is only one partial mitochondrial 16S rRNA gene sequence in the GenBank database (i.e. accession HM211198). Thus, the determination of the complete mitochondrial genome will be useful for the identification of potential DNA markers for aquaculture, biodiversity and population structure studies, and to obtain phylogenetic insights.
Specimens were collected from El Encanto beach (8°37'05.6"S 78°45'29.0"W, Virú, La Libertad, Peru, n = 3) and from an aquaculture facility Pacific Deep Frozen (Huarmey, Ancash, Peru, n = 4). Then, seven male specimens were transferred to the Laboratorio of Genética, Fisiología y Reproducción (Universidad Nacional del Santa, Ancash, Peru) and registered under the voucher numbers PA-CH1, PA-CH2, PA-CH3, PA-PDF1M, PA-PDF5M, PA-PDF6M, and PA-PDF12M. Each total genomic DNA was isolated from 10 mg of muscle tissue using an automated iPrep purification system (Invitrogen, Life Technologies; Thermo Fisher Scientific Inc., Waltham, MA, USA). DNA libraries were prepared using Nextera DNA Flex Library Prep (Illumina, San Diego, CA, USA) and paired-end (150 bp each) sequencing on a NextSeq 500 platform (Illumina, San Diego, CA, USA). Subsequently, the clean paired-end reads were assembled following the Norgal v1.0 tool pipeline (Al-Nakeeb et al. Citation2017), with minor modifications. For example, for the removal of sequencing adapters, Trimmomatic v0.36 (Bolger et al. Citation2014) was used instead of Adapter removal from Norgal. Trimming parameters were ILLUMINACLIP:illumina_all_adapters.fa:2:30:10 LEADING:5 TRAILING:5 SLIDINGWINDOW:5:25 MINLEN:50. Also, the maximum k-mer for megahit assembly was set to 100 instead of 105.
The complete circular mitogenome of P. adspersus presented 17,060 bp in size (GenBank accession no. MW288827), showing a nucleotide composition of: A 27.5%, C 29.5%, G 17.1%, and T 25.9%. Its mitogenome contains 13 protein-coding genes (PCGs) which were identified by an ORF finder analysis at NCBI (https://www.ncbi.nlm.nih.gov/orffinder/), and two ribosomal RNA genes were identified by sequence alignments and compared to several related species. Moreover, 22 transfer RNA genes were identified with tRNAscan-SE (Chan and Lowe Citation2019); and a control region (D-loop) of 1368 nt was also identified. All PCGs use the typical ATG start codon, except for the COI gene which utilizes the alternative GTG start codon. Six PCGs ended with the complete termination codons TAA (ND1, ATPase 8, ND4L, ND5, and ND6) or TAG (COI), whereas the remaining six PCGs used the incomplete stop codons TA– (ND2, ATPase 6, and COIII) or T–– (COII, ND3, ND4, and Cytb). These incomplete stop codons are presumably completed to TAA by posttranscriptional polyadenylation (Ojala et al. Citation1981). Except for ND6 gene and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro), all other genes were located on the heavy strand (H-strand).
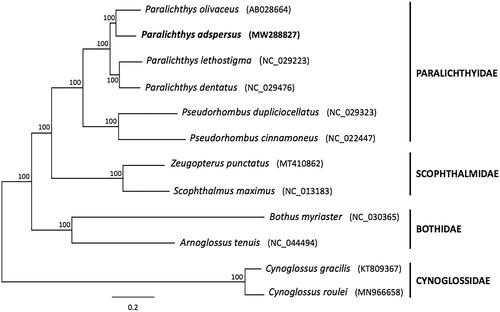
Finally, to infer the phylogenetic position of P. adspersus, a Bayesian analysis was performed based on the concatenated sequences of 12 PCGs (ND6 was excluded) from 12 pleuronectiform species using MrBayes 3.2 (Ronquist and Huelsenbeck Citation2003). Substitution saturation in single codon positions from each PCGs was analyzed using DAMBE5 (Xia Citation2013). Substitution model selection for all individual genes (12 PCGs) was performed using the software jModelTest 2 (Darriba et al. Citation2012). The best-fit substitution model for each partition was selected according to the Bayesian Information Criterion (BIC). As shown in the phylogenetic tree (), six Paralichthyidae species included in the analysis constituted a monophyletic group, with P. adspersus gathered into a single subclade with P. olivaceus, while the clade formed by both species from the genus Pseudorhombus was placed at the most basal position of Paralichthyidae, reinforcing the data obtained from the present analysis.
Figure 1. Bayesian phylogenetic tree inferred from 12 concatenated mitochondrial protein-coding genes (ND6 was excluded) of pleuronectiform species. The sequence matrix (10,684 nt) used in the phylogenetic analyses consisted of unambiguously aligned regions of the first, second, and third codon positions (COI and ATPase 8), all other PCGs included only first and second codon positions. The GTR + I+G model was estimated as the best-fit substitution model for genes ATPase6, COII, ND1, ND2, and ND5; HKY + G model for genes ATPase8 and COI; HKY + I+G model for genes COIII, Cytb, and ND4; and GTR + G model for genes ND3 and ND4L. The position of Paralichthys adspersus (whose mitogenome was determined in this study) is shown in bold. Posterior probabilities at correspondent nodes are shown in percentages. GenBank accession numbers for each species are shown in parentheses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in [GenBank reference number MW288827, https://www.ncbi.nlm.nih.gov/nuccore/MW288827].
Additional information
Funding
References
- Al-Nakeeb K, Petersen TN, Sicheritz-Pontén T. 2017. Norgal: extraction and de novo assembly of mitochondrial DNA from whole-genome sequencing data. BMC Bioinf. 18(1):1–7.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Chirichigno N, Cornejo RM. 2001. Catálogo comentado de los peces marinos del Perú. Lima: Instituto del Mar del Perú; p. 314.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 30(7):1720–1728.
