Abstract
Verbena officinalis has a long history as a source plant in traditional Chinese medicine. This study adopted next-generation sequencing technology in order to determine complete chloroplast genome of V. officinalis. The results of this investigation showed the chloroplast genome of V. officinalis was 153,286 bp in length, including a pair of inverted repeat (IR) regions (each 25,825 bp), separated by a large single-copy region (LSC) of 84,316 bp and a small single-copy region (SSC) of 17,320 bp, and the overall GC contents of the chloroplast genome was 39.04%. Additionally, we annotated 83 genes, including 48 protein-coding genes, 31 tRNA genes, and 4 rRNA genes. By creating the phylogenetic tree, relationship between V. officinalis and relevant species was discussed, and the result proved that V. officinalis was closely related to Avicennia marina. The findings of the study will serve as a stepping stone for follow-up researches regarding its chloroplast genome.
Verbena officinalis is a perennial plant species, which has been widely used as a herbal medicine in China for centuries. Besides, both the Chinese Pharmacopeia and the European Pharmacopeia have included V. officinalis (Kubica et al. Citation2020). It is distributed from temperate to tropical regions all over the world. V. officinalis grows on roadsides, hillsides, streams or forests at low to high altitudes (Waheed et al. Citation2016). At present, it is also gradually beginning to be used as an ornamental plant because of its purple flowers. The previous study showed that extracts from the leaves of V. officinalis had anti-inflammatory (Deepak and Handa Citation2000), antioxidant and antifungal activities (Casanova et al. Citation2008). However, studies on V. officinalis were mainly in the fields of population biology, ecology, physiology, and stress resistance. The research on characterization of the chloroplast genome of Verbena officinalis is still in infancy, and further investigation is needed.
The genomic DNA of V. officinalis was extracted from its freshly-picked leaves which were collected in Heifei, Anhui, China (N31°56′17″; E117°23′25″). The plant specimens were kept in the Herbarium of Anhui University of Chinese Medicine with the voucher number 200809AH001. We took an approach to extract genomic DNA by using CTAB method (Borges et al. Citation2012). In order to obtain the sequence raw data, we entrusted Genewiz Co. Ltd. (Suzhou, China) to conduct next-generation sequencing. After discarding and trimming reads with low quality, the clean data were assembled and gapfilled by Trinity and SSPACE (version 3.0). Based on the assembly results, the coding gene, rRNA, tRNA and other ncRNAs were predicted, and then the predicted genes were analyzed by functional annotation of KEGG database (Ogata et al. Citation1999).
The complete chloroplast genome sequence of V. officinalis was uploaded to GenBank with the accession number MW348926. The final result showed that genomic sequence length was 153,286 bp, which consisted of a pair of inverted repeat (IR) regions (each 25,825 bp), separated by a large single-copy region (LSC) of 84,316 bp and a small single-copy region (SSC) of 17,320 bp. The GC contents in the chloroplast genome were found to be 39.04%. The chloroplast genome of Verbena officinalis contained 83 genes, including 48 protein-coding genes, 31 tRNA genes, and 4 rRNA genes.
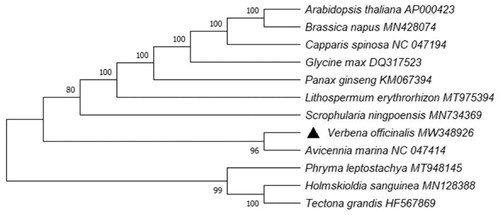
The phylogenetic trees help in acquiring the evolutionary history of Verbena officinalis. Therefore, to study evolutionary information, we established phylogenetic tree by using MEGA X (Kumar et al. Citation2018). The analysis was based on complete chloroplast genome sequence of V. officinalis, along with other 11 relevant species downloaded from GenBank. In the process, the chloroplast genome sequences were aligned with ClustalW. Subsequently, we chose Bootstrap method to conduct phylogeny test (1000 replicates), with a combining method of Tamura-Nei substitution model (Tamura and Nei Citation1993) to generate maximum likelihood (ML) tree (Kumar et al. Citation2016). The finding of this study suggested that V. officinalis was closely related to Avicennia marina which is from the same family of Verbenaceae ().
Disclosure statement
No potential conflict of interest was reported by author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW348926. The associated BioProject, Bio-Sample and SRA numbers are PRJNA688015, SAMN17167431, and SRR13309663 respectively.
Additional information
Funding
References
- Borges DB, Amorim MB, Waldschmidt AM, Mariano-Neto E, Vivas CV, Pereira DG. 2012. Optimization of DNA extraction from fresh leaf tissues of Melanoxylon brauna (Fabaceae)). Genet Mol Res. 11(2):1586–1591.
- Casanova E, García-Mina JM, Calvo MI. 2008. Antioxidant and antifungal activity of Verbena officinalis L. Leaves. Plant Foods Hum Nutr. 63(3):93–97.
- Deepak M, Handa SS. 2000. Antiinflammatory activity and chemical composition of extracts of Verbena officinalis. Phytother Res. 14(6):463–465.
- Kubica P, Szopa A, Dominiak J, Luczkiewicz M, Ekiert H. 2020. Verbena officinalis (Common Vervain) – a review on the investigations of this medicinally important plant species. Planta Med. 86(17):1241–1257.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kumar S, Stecher G, and, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27(1):29–34.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526.
- Waheed KA, Arif-Ullah K, Touqeer A. 2016. Anticonvulsant, anxiolytic, and sedative activities of Verbena officinalis. Front Pharmacol. 7:499.