Abstract
Oxytropis glabra DC. is a perennial poisonous plant to livestock belonging to the genus Oxytropis, Leguminosae, mainly distributed in Northwestern China. As a poisonous grass, this species protects plant diversity in degraded grasslands by sheltering adjacent plants. In this study, the complete chloroplast genome with a total size of 122,094 bp was reported. Our annotations showed that the chloroplast genome contains 109 genes, including 76 protein-coding genes, 29 tRNA genes, and four rRNA genes. This work presents complete chloroplast genome information, which will be valuable for studying the evolution and genetic diversity of O. glabra.
Oxytropis glabra DC. (Leguminosae) is a perennial poisonous plant to livestock belonging to Leguminosae, mainly distributed in Northwestern China. As a common poisonous grass, O. glabra protects plant diversity in degraded grasslands by sheltering adjacent plants. In addition, some species of Oxytropis were reported to have high flavonoids, such as Oxytropis falcata Bunge. The whole herb of Oxytropis falcata has a variety of pharmacological activities, including anti-inflammatory and antioxidant drugs (Wang et al. Citation2010; Yang et al. Citation2010). In order to reveal the phylogeny of O. glabra, we sequenced the genome, assembled and annotated the complete chloroplast genome.
In this study, the materials of O. glabra were collected from Akqi County, Xinjiang province of China (78.675°E, 41.006°N, 1837 m above sea level). The voucher specimen (TD-00572, Oxytropis glabra DC.) was stored in the herbarium of Tarim University. The total genomic DNA from leaves was extracted using CTAB method (Doyle and Doyle Citation1987) and sequenced using the Illumina NovaSeq platform at Majorbio Company (Shanghai, China). First, the clean data were quality-controlled by using FastQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The whole chloroplast genome was assembled using GetOrganelle v1.7.3 (Jin et al. Citation2020). Then, to check the accuracy of assembly results, the slimmed assembly graph and selected target assembly graph can be visualized by Bandage v0.8.1 (https://github.com/rrwick/Bandage/releases/tag/v0.8.1) to assess the completeness of the final graph. Finally, the final assembly result is obtained. Gene annotation was performed using CPGAVAS2 (http://47.96.249.172:16019/analyzer/annotate) (Shi et al. Citation2019) and PGA (https://github.com/quxiaojian/PGA) (Qu et al. Citation2019). The differential annotations of protein-coding sequences were confirmed using BLASTx in NCBI. We obtained a complete chloroplast genome of 122,094 bp (MW349014) that lost an IR region and included average GC content of 34.3%. Most chloroplast genome are characterized by a quadripartite structure, which include two copies of an inverted repeat (IR) separating the large (LSC) and small (SSC) single copy regions. But some tribes among legumes have a common phenomenon that losing one copy of the IR in the chloroplast genome, such as Carmichaelieae, Cicereae, Hedysareae, Trifolieae, Fabeae, Galegeae, and three genera of Millettieae (Palmer and Thompson Citation1982; Lavin et al. Citation1990; Liston Citation1995; Jansen et al. Citation2008). The phenomenon may be a special feature of legumes in the evolutionary process. In this study, we showed that the complete chloroplast genomes encoded 109 functional genes, containing 76 protein-coding genes, 29 tRNA genes, and four rRNA genes.
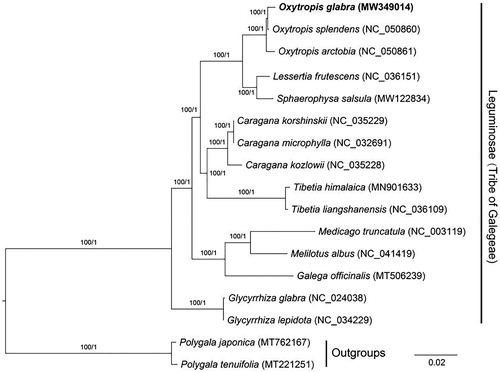
To reveal the phylogenetic relationship of O. glabra within Leguminosae, additional 14 species from Leguminosae were selected to study. With the Polygala japonica and Polygala tenuifolia as the outgroups, the phylogenetic trees were built from the 76 protein-coding gene matrixes by maximum-likelihood (ML) and Bayesian inference (BI) (). The ML tree was generated using IQ-TREE v2.1.2 (Nguyen et al. Citation2015) based on the best model of TVM + F+R2 and 1000 bootstrap replicates, and BI analysis was performed in MrBayes v3.2.7 (Ronquist et al. Citation2012). This result showed that the analyzed O. glabra was clustered with O. splendens and O. arctobia, all of which showed closer to the species of Lessertia frutescens and Sphaerophysa salsula.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW349014. The associated “BioProject”, “SRA”, and “Bio-Sample” numbers are PRJNA686236, SRR13275092, and SAMN17109568 respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell H. 2008. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol Phylogenet Evol. 48(3):1204–1217.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Lavin M, Doyle JJ, Palmer JD. 1990. Evolutionary significance of the loss of the chloroplast–DNA inverted repeat in the Leguminosae subfamily Papilionoidae. Evolution. 44(2):390–402.
- Liston A. 1995. Use of the polymerase chain reaction to survey for the loss of the inverted repeat in the legume chloroplast genome. In: Crisp M; Doyle JJ, editors. Advances in legume systematics 7: phylogeny. Kew: Royal Botanic Gardens; p. 31–40.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Palmer JD, Thompson WF. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 29(2):537–550.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Wang D, Tang W, Yang GM, Cai BC. 2010. Anti-inflammatory, antioxidant and cytotoxic activities of flavonoids from Oxytropis falcata bunge. Chin J Nat Med. 8(6):461–465.
- Yang GM, Wang D, Tang W, Chen X, Fan LQ, Zhang FF, Yang H, Cai BC. 2010. Anti-inflammatory and antioxidant activities of Oxytropis falcata fractions and its possible anti-inflammatory mechanism. Chin J Nat Med. 8(4):285–292.