Abstract
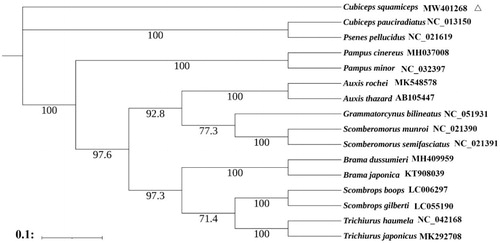
The Indian driftfish (Cubiceps squamiceps) is one of the most important commercial fish species in China, Japan and India. The complete mitogenome of Cubiceps squamiceps was determined in this study. The assembled mitogenome was 16,507 bp and consisted of 13 protein-coding genes, 22 tRNAs, two rRNAs, and a control region. Nucleotide composition of the complete mitogenome was 27.5% A, 28.5% C, 17.5% G, and 26.5% T, with an A + T bias of 53.9%. The maximum-likelihood tree based on 13 protein-coding genes showed that Cubiceps pauciradiatus and Psenes pellucidus were the closest to C. squamiceps.
Cubiceps squamiceps belongs to genus Cubiceps, family Nomeidae, order Stromateoidei. It is widely distributed in the coast of India-Western Pacific (Zhao et al. Citation2016). Previous studies on C. squamiceps were mainly related to its distribution and biological characteristics. For example, Kim et al. (Citation1988) found five new fish species collected from the southern coastal waters of Korea, one of which was C. squamiceps. Guang et al. (Citation2017) determined the concentrations of heavy metals (Cd, Pb, Cr, Ni, Cu and Zn) in C. squamiceps and found that concentrations of heavy metals in C. squamiceps were below their acceptable daily upper limit, suggesting human consumption of these wild fish species may be safe. The complete mitochondrial genome was considered as a useful tool for species identification, phylogeny and evolution (Zhang and Georges Citation2014). There were a few researches on evolution and phylogenetic analysis of this fish. In this study, we described the complete mitochondrial genome of C. squamiceps and explored the phylogenetic relationship within Scombriformes, which can provide an important dataset for a better understanding of the mitogenomic diversities.
Firstly, samples of C. squamiceps were collected from the East China Sea (N 27°49′52.12″, E 121°10′31.21″) and stored in a laboratory of Zhejiang Ocean University with accession number 20200513bj19. The email of the people in charge of the collection is [email protected]. Total genomic DNA was extracted using standard phenol-chloroform extraction procedure (Sambrook and Russell Citation2006). Subsequently, based on the existing complete mitochondrial gene of Psenes pellucidus (NC_021619), 19 pairs of primers were designed. The samples were amplified by PCR and sequenced using Sanger sequencing technology. The complete mitochondrial genome was annotated using Sequin version 15.10 (http://www.ncbi.nlm.nih.gov/Sequin) and tRNAscan-SE version 2.0 (http://trna.ucsc.edu/tRNAscan-SE/; Lowe and Eddy Citation1997). The mitochondrial genome of C. squamiceps has been stored in the GenBank with accession number MW401268. The complete mitogenome of the C. squamiceps was 16,507 bp in length including two rRNA genes, 13 protein-coding genes, 22 tRNA genes and a control region, which was similar to other typical vertebrate mitochondria (Boore Citation1999). Compared with the sequence stored in GenBank with Acc Nos. KT_361215 and NC_029845, we had provided a new sequence with a difference of 3 bases. The different sequence numbers of the same species may be caused by the different sampling sites. The overall base composition was 27.5% (A), 26.5% (T), 28.5% (C), and 17.5% (G) with a slight A + T bias (53.9%), which was similar to other typical Nomeidae fish mitogenomes (Wei et al. Citation2014). 12 genes started with ATG while only COI started with GTG among the 13 protein-coding genes. Seven genes shared the termination codon TAA including ND1, COI, ATP8, ATP6, COIII, ND4L and ND5, and the gene with TAG as the stop codon was ND6. The remaining protein-coding genes with incomplete termination codons were COII (T–), ND2 (TA-), ND3 (T–), ND4 (T–), and Cytb (T–). Besides, nine genes were coded on the L-strand including tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, tRNA-Pro and the ND6 gene, the others were encoded on the H-strand which was consistent with other fish mitochondrial genes (Yukai et al. Citation2019).
Maximum-Likelihood phylogeny was constructed based on 13 protein-coding genes of C. squamiceps and other 15 Scombriformes using the software PhyML 3.0 (Guindon et al. Citation2010). The most suitable nucleotide sequence model GTR + I + G was selected through MrModeltest 2.3 based on the Akaike Information Criteria (AIC) (Yamaoka et al. Citation1978). The phylogenetic analysis showed that Cubiceps pauciradiatus and Psenes pellucidus were the closest to C. squamiceps which was consistent with the result of Zhao et al. (Citation2016). The complete mitochondrial genome sequence of the C. squamiceps can provide an important dataset for a better understanding of the mitogenomic diversities, evolution in C. squamiceps and facilitating the identification of the species ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statements
Mitochondrial genome sequence can be accessed via accession number MW401268 (https://www.ncbi.nlm.nih.gov/nuccore/MW401268) in the NCBI GenBank.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Guang GY, Lin Q, Hui HH, Gen WL, Jia NJ, Yan DF. 2017. Heavy metals in fish tissues/stomach contents in four marine wild commercially valuable fish species from the western continental shelf of South China Sea. Mar Pollut Bull. 114(2):1125–1129.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Kim KH, Kim YU, Kim YS. 1988. Five species of fish new to Korean Waters. Korean J Fish Aquat Sci. 21:105–112.
- Lowe T, Eddy S. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Sambrook J, Russell DW. 2006. Purification of nucleic acids by extraction with phenol:chloroform. Csh Protoc. 2006(1):pdb.prot4455.
- Wei T, Zhang B, Sun Y. 2014. Structure comparison of control region in Stromateoidei and complete mitochondrial genome of the bluefin driftfish Psenes pellucidus (Perciformes, Nomeidae). Mitochondrial DNA. 25(5):355–356.
- Yamaoka K, Nakagawa T, Uno T. 1978. Application of Akaike’s information criterion (AIC) in the evaluation of linear pharmacokinetic equations. J Pharmacokinet Biopharm. 6(2):165–175.
- Yukai Y, Xiaolin H, Heizhao L, Tao L, Wei Y, Zhong H. 2019. The complete mitochondrial genome of Chaetodon wiebeli (Chaetodontiformes, Chaetodontidae). Mitochondrial DNA B Resour. 4(2):3145–3146.
- Zhang X, Georges A. 2014. A complete mitochondrial genome sequence for the Australian turtle, Chelodina longicollis, obtained using 454-pyrosequencing. Conserv Genet Resour. 6:555–557.
- Zhao M, Ma H, Ma C, Zhang H, Zhang X, Meng Y, Wei H, Chen F, Ma L. 2016. The complete mitochondrial genome and gene organization of Cubiceps squamiceps (Perciformes: nomeidae) with phylogenetic consideration. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4296–4297.