Abstract
Catalpa fargesii Bur. is endemic to China. Its complete chloroplast genome sequence was firstly reported in this study. The whole chloroplast genome of this species was 157765 bp in length including a pair of inverted repeat (IR, 30252 bp) regions separated by a small single copy (SSC, 12662 bp) and a large single copy (LSC, 84599 bp). The genome consisted of 134 genes, including 89 protein-coding genes, 8 rRNA and 37 tRNA genes. The phylogenetic analysis strongly supported that C. fargesii was closely related to C. fargesii f. duclouxii and C. bungei.
Catalpa fargesii Bur. is a deciduous tree, up to 25 m. It is endemic to China and mainly distributed in northwestern China. This species is an important economic tree species used as valuable timer for furniture, appliance and building because of its high density and hardness (Ma et al. Citation2021). C. fargesii also can be used horticulturally as garden and street trees for its large heart-shaped leaves and showy flowers (Ma et al. Citation2021). Besides, it can resist extreme drought and coldness. In the recent taxonomic revision of the genus Catalpa, Catalpa fargesii and C. fargesii var. duclouxii were treated as a single species, C. catalpa (Olsen and Kirkbride Citation2017). However, we recognized them to be different in several characters, including indumentum on young branches and leaves, leaf baldes, inflorescence, corolla, anthers. For instance, stellate or dendroid hairs of C. fargesii could differ it from C. fargesii var. duclouxii and C. bungei.
The sample of C. fargesii was collected from Maiji County, Gansu province, China (E 105.93920, N 34.44944). The voucher specimen (Ma WJ, HQ-1) was deposited at the Herbarium of Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China (CAF) and DNA sample was properly stored at Key Laboratory of Tree Breeding and Cultivation of Forestry and Grassland Administration; Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China. Total genomic DNA was extracted from the leaves dried with silica gel using the modified CTAB method of Doyle and Doyle (Citation1987). For short-read sequencing, an ∼150 bp insert size pair-end library was constructed and sequenced using the Illumina HiSeq 2000 platform in the Sangon Biotech (Shanghai, China). A total of 5.5 Gb raw data were generated. By using SPAdes 3.13.0 (Bankevich et al. Citation2012) and Geneious 9.0.5 (http://www.geneious.com/), all contigs of the chloroplast genome sequence were spliced and assembled. Then, the Geneious 9.0.5 also was applied to annotate the complete chloroplast genome. The whole chloroplast genome of Catalpa fargesii f. duclouxii (MT783420) was used as a reference sequence. The complete chloroplast genome sequence has been deposited in GenBank with an accession number MW338733.
The circular chloroplast DNA of C. fargesii is 157,765 bp in length, and the average GC content was 37.43%. It consists of a typical quadripartite structure with two inverted repeats (IRs) of 30252 bp separated by a large single copy (LSC) of 84599 bp and a small single copy (SSC) of 12662 bp. The complete chloroplast genome of C. fargesii contains 134 unique genes, including 89 protein-coding genes, 37 tRNA genes and 8 rRNA genes. In these genes, 7 genes (2 tRNA genes and 5 protein-coding genes) contain one intron, whereas three genes (ycf3, rpl2, and ndhB) contain double introns.
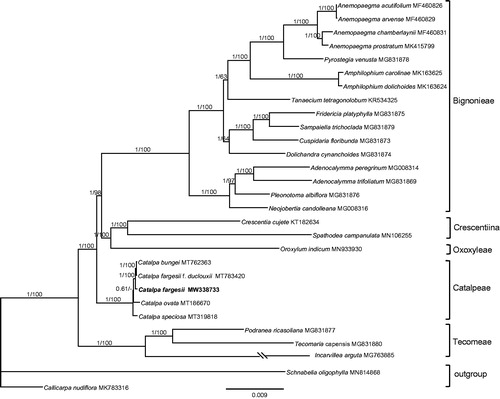
To confirm the phylogenetic position of C. fargesii within the family of Bignoniaceae, additional 27 complete chloroplast genomes of Bignoniaceae were obtained from GenBank, and two species, Schnabelia oligophylla and Callicarpa nudiflora were used as the outgroups (). The 29 complete chloroplast sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). Phylogenetic analysis was conducted by using PAUP 4.0b10 (Swofford Citation2003) with 1000 bootstrap replicates and MrBayes 3.2.6 (Ronquist and Huelsenbeck Citation2003). The nucleotide substitution model was selected by the Akaike information criterion (AIC) in Modeltest 3.7 (Posada and Crandall Citation1998). For BI analysis, four chains of the Markov Chain Monte Carlo (MCMC) were run for 5,000,000 generations starting with a random tree, sampling one tree every 1000 generations. Majority-rule (>50%) consensus trees were generated after removing a 25% burn-in. Phylogenetic analysis results strongly supported that C. fargesii was closely related to a clade including C. fargesii f. duclouxii and C. bungei (MP-BS = 100, BI-PP = 1.00) (). The phylogenetic relationship of Bignoniaceae recovered from this study is congruent with that of Olmstead et al. (Citation2009).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the finding of this study is available in GenBank at https://www.ncbi.nlm.nih.gov/ with an accession number MW338733. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA715373, SAMN18347870, and SRR14027477, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Ma WJ, Xiao Y, Li Y, Hu P, Wang Z, Yang GJ, Wang JH. 2021. Overexpression of CfPIP1-1, CfPIP1-2, and CfPIP1-4 genes of Catalpa fargesii in transgenic Arabidopsis thaliana under drought stress. J for Res. 32(1):285–296.
- Olmstead RG, Zjhra ML, Lohmann LG, Grose SO, Eckert AJ. 2009. A molecular phylogeny and classification of Bignoniaceae. Am J Bot. 96(9):1731–1743.
- Olsen RT, Kirkbride JH. 2017. Taxonomic revision of the genus Catalpa (Bignoniaceae). Brittonia. 69(3):387–421.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Swofford D. 2003. PAUP*-phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland (MA): Sinauer associates.