Abstract
Monolepta hieroglyphica (Motschulsky, 1858) is a major pest of potato, maize, cotton and sorghum in China. In this study, we sequenced and analyzed the complete mitochondrial genome (mitogenome) of M. hieroglyphica. This mitogenome was 15,761 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs) and two ribosomal RNA unit genes (rRNAs). Gene order was conserved and identical to most other previously sequenced Galerucinae Most PCGs of M. hieroglyphica have the conventional start codons ATN (six ATT, five ATG and one ATC), with the exception of nad1 (TTG). Except for three genes (cox1, nad4 and nad5) end with the incomplete stop codon T––, all other PCGs terminated with the stop codon TAA or TAG. The whole mitogenome exhibited heavy AT nucleotide bias (80.0%). Phylogenetic analysis positioned M. hieroglyphica in a well-supported clade within the subfamily Galerucinae with Monolepta occifluvis, Monolepta sp. and Paleosepharia posticata. These results provided an important basis for further studies on mitochondrial genome and phylogenetics of Galerucinae.
The family Chrysomelidae, commonly known as leaf beetles, includes over 37,000 species in more than 2000 genera, making it one of the largest and most commonly encountered of all beetle families (Thormann et al. Citation2016). Most leaf beetles are small to medium-sized, with the antennae notably shorter than head, thorax and abdomen. The Galerucinae is the largest subfamily of the Chrysomelidae and includes about 14,500 described species occurring worldwide (Nie et al. Citation2018). Adult and larval leaf beetles within Galerucinae feed on all sorts of plant tissue, and all species are fully herbivorous (Nie et al. Citation2018; Zheng et al. Citation2020). Many of them are serious pests of cultivated plants, for example the widespread leaf beetle Monolepta hieroglyphica (Motschulsky, 1858), which can damage potato, maize, cotton and sorghum. Herein, we sequenced and analyzed its mitogenome.
Five male adults of M. hieroglyphica were collected from Tianshui City, Gansu Province, China (34°27′N, 105°43′E, July 2019) and were stored in Entomological Museum of Qinghai University (Accession number QHU-EMMH02). Total genomic DNA was extracted from muscle tissues of the thorax using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany). A pair-end sequence library was constructed and sequenced using Illumina HiSeq 2500 platform (Illumina, San Diego, CA), with 150 bp pair-end sequencing method. A total of 20.4 million reads were generated and had been deposited in the NCBI Sequence Read Archive (SRA) with accession number SRR13319950. Raw reads were assembled using MITObim v 1.7 (Hahn et al. Citation2013). By comparison with the homologous sequences of other Galerucinae species from GenBank, the mitogenome of M. hieroglyphica was annotated using software GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand).
The complete mitogenome of M. hieroglyphica is 15,761 bp in length (GenBank accession no. MW419951), and contains the typical set of 13 protein-coding, two rRNA and 22 tRNA genes, and one non-coding AT-rich region. Gene order was conserved and identical to most other previously sequenced Chrysomelidae (Nie et al. Citation2018; Song et al. Citation2018; Long et al. Citation2019; Wang et al. Citation2020). The nucleotide composition of the mitogenome is 80.0% A + T content (A 41.1%, T 38.9%, C 11.9%, G 8.1%). Four PCGs (nad4, nad4l, nad5 and nad1) were encoded by the minority strand (N-strand) while the other nine were located on the majority strand (J-strand). The 22 tRNA genes vary from 61 bp (trnC and trnL1) to 71 bp (trnV). Two rRNA genes (rrnL and rrnS) locate at trnL1/trnV and trnV/control region, respectively. The lengths of rrnL and rrnS in M. hieroglyphica are 1273 and 740 bp respectively, with AT contents of 83.2% and 84.2%, respectively. Most PCGs of M. hieroglyphica have the conventional start codons ATN (six ATT, five ATG and one ATC), with the exception of nad1 (TTG). Except for three genes (cox1, nad4 and nad5) end with the incomplete stop codon T––, all other PCGs terminated with the stop codon TAA or TAG. Genetic biodiversity of this worldwide pest in China is still unclear, which can be resolved by sequencing more genetic data of different geographical populations. Compared with the mitogenome of M. hieroglyphica which already existed in Genbank (MT192098, collected from Zhoukou City, Henan Province, China) (Li et al. Citation2020), gene rearrangement, base composition and other general features of this two mitochondrial genomes were similar to each other. However, 96.3% percentage of pairwise identity was shown based on the MAFFT alignment, indicating the obvious nucleotide diversity of M. hieroglyphica between this two geographical populations, this may be caused by the geographical isolation and can provide important genetic information for futher population genetic structure analysis of M. hieroglyphica.
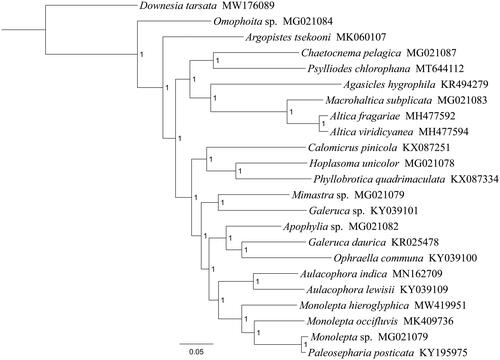
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 23 Coleoptera species. Alignments of individual genes were concatenated using SequenceMatrix 1.7.8 (Vaidya et al. Citation2011). Optimal nucleotide substitution models and partition strategies were chosen by PartitionFinder v1.1.1 (Lanfear et al. Citation2012). Following the partition schemes suggested by PartitionFinder, phylogenetic tree was constructed using MrBayes 3.2.6 (Ronquist and Huelsenbeck Citation2003). Two simultaneous runs with four independent Markov chains were run for five million generations and trees were sampled every 1000th generation. After the average standard deviation of split frequencies fell below 0.01, the first 25% of samples were discarded as burn-in and the remaining trees were used to generate a consensus tree and calculate the posterior probabilities (PP). Phylogenetic analysis positioned M. hieroglyphica in a well supported clade within the subfamily Galerucinae with Monolepta occifluvis, Monolepta sp. and Paleosepharia posticata (), indicating genus Monolepta had a close relationship with Paleosepharia. The monophyly of Monolepta could not be confirmed by this phylogenetic tree. These results provided an important basis for further studies on mitochondrial genome and phylogenetics of Galerucinae.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MW419951. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA688388, SRR13319950, and SAMN17174216 respectively.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Li JH, Shi YX, Song L, Zhang Y, Zhang HF. 2020. The complete mitochondrial genome of an important agricultural pest Monolepta hieroglyphica (Coleoptera: Chrysomelidae: Galerucinae). Mitochondr DNA B. 5(2):1820–1821.
- Long C, Liu P, Guo Q, Xu J, Dai X. 2019. Complete mitochondrial genome of a leaf-mining beetle, Argopistes tsekooni (Coleoptera: Chrysomelidae). Mitochondr DNA B. 4(1):418–419.
- Nie RE, Breeschoten T, Timmermans MJ, Nadein K, Xue HJ, Bai M, Huang Y, Yang XK, Vogler AP. 2018. The phylogeny of Galerucinae (Coleoptera: Chrysomelidae) and the performance of mitochondrial genomes in phylogenetic inference compared to nuclear rRNA genes. Cladistics. 34(2):113–130.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Song N, Yin X, Zhao X, Chen J, Yin J. 2018. Reconstruction of mitogenomes by NGS and phylogenetic implications for leaf beetles. Mitochondrial DNA A DNA Mapp Seq Anal. 29(7):1041–1050.
- Thormann B, Ahrens D, Armijos D, Peters M, Wagner T, Wägele JW. 2016. Exploring the leaf beetle fauna (Coleoptera: Chrysomelidae) of an ecuadorian mountain forest using DNA barcoding. PLoS One. 11(2):e0148268.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Wang H, Bai Y, Li G, Luo J, Li C. 2020. Characterization of the complete mitochondrial genome of Aulacophora indica (Insecta: Coleoptera: Chrysomeloidea) from Zhijiang. Mitochondr DNA B. 5(2):1459–1460.
- Zheng F, Jiang H, Jia J, Wang R, Zhang Z, Xu H. 2020. Effect of dimethoate in controlling Monolepta hieroglyphica (Motschulsky) and its distribution in maize by drip irrigation. Pest Manag Sci. 76(4):1523–1530.