Abstract
Rosa sterilis is an economically and important fruit that is extensively grown in Southwestern China. In this study, we determined the complete chloroplast genome of R. sterilis using high-throughput Illumina sequencing. The chloroplast genome of R. sterilis is 156,561 bp in size, containing a large single-copy region (LSC)(85,701 bp), a small single-copy region (SSC) (18,746 bp), and a pair of inverted repeat (IR) regions (each one of 26,057 bp). The overall GC content of the chloroplast genome is 37.23%, while the corresponding values of GC contents of the LSC, SSC, and IR regions are 35.20%, 31.37%, and 42.70%, respectively. The chloroplast genome of R. sterilis contains 130 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic maximum-likelihood tree revealed that Rosa chinensis or Rosa chinensis var. spontanea is the closest related to R. sterilis in the phylogenetic relationship. This complete chloroplast genome can be further used for genomic studies, evolutionary analyses, and genetic engineering studies of the family Rosaceae.
Rosa sterilis, belonging to the Rosaceae family, is a perennial shrub and originates from the Karst areas of Guizhou Province (Liu et al. Citation2016). The fruits of R. sterilis have been widely consumed due to good flavor and bioactivities compounds, such as triterpenes, amino acids, flavonoids, and other phenylpropanoid derivatives (Luo et al. Citation2017). The aim of this study was to sequence, assemble, and characterized the complete chloroplast genome of R. sterilis to learn more about genetic knowledge. R. sterilis was planted under natural conditions in Guizhou Normal University, Guizhou province (Guiyang, China 26°42.408′ N; 106°67.353′ E). The leaves of R. sterilis were collected and deposited at the herbarium of Guizhou Normal University (GZNU-PRR-ML-01). The total genomic DNA of R. sterilis was extracted using the optimized CTAB method and sequenced using the high-throughput Illumina NovaSeq6000 Sequencing Platform System (Illumina Co., San Diego, CA).
Quality control was performed to remove low-quality reads and adapters using the FastQC (Andrews Citation2015). A total of 980,062 clean reads were obtained and the average value of Q30 is 90.88%, indicating the good quality for further analysis. The chloroplast genome was assembled using the SPAdes version 3.5.0 (http://cab.spbu.ru/software/spades/) and annotated using the CpGAVAS (Liu et al. Citation2012; Lapidus et al. Citation2014). The tRNA genes were further identified using ARWEN (http://mbio-serv2.mbioekol.lu.se/ARWEN/) and tRNAscan-SE (Lowe and Chan Citation2016; Laslett and Canbäck Citation2008). The physical map of the chloroplast genome of R. sterilis was generated using Organellar Genome DRAW. Clean reads were obtained with 150-bp paired-end reads. The complete chloroplast genome sequence together with corresponding annotations has been submitted to Genbank under the accession number of MW007387.
The chloroplast genome of R. sterilis is a circle molecular genome with the length of 156,561 bp, containing a large single-copy region (LSC) of 85,701 bp, a small single-copy region (SSC) of 18,746 bp, and a pair of inverted repeat (IR) regions of 26,057 bp in each one. The overall nucleotide consists of A (48,512 bp), T (49,745 bp), C (29,638 bp), and G (28,657 bp), with the total GC content is 37.23%, similar to other species in the Rosaceae (Huang and Cronk Citation2015). The largest GC content ratio was obtained in the IR regions (42.70%). The values of GC content ratio in the LSC and SSC region were 35.20% and 31.37% because the tRNA and rRNA genes extensively have fewer AT nucleotides (He et al. Citation2020). The chloroplast genome of R. sterilis comprised 130 genes, including 84 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. Most genes for photosynthesis were localized in the LSC and SSC regions. In the IR regions, we determined seven protein-coding genes (ycf2, ndhB, ycf1, rps12, rpl2, rpl23, and rps7,), seven kinds of tRNA (tRNA-CAT, tRNA-CAA, tRNA-GAC, tRNA-GAT, tRNA-TGC, tRNA-ACG, and tRNA-GTT), and four kinds of rRNA (rrn16 S, rrn23 S, rrn4.5S, and rrn5S).
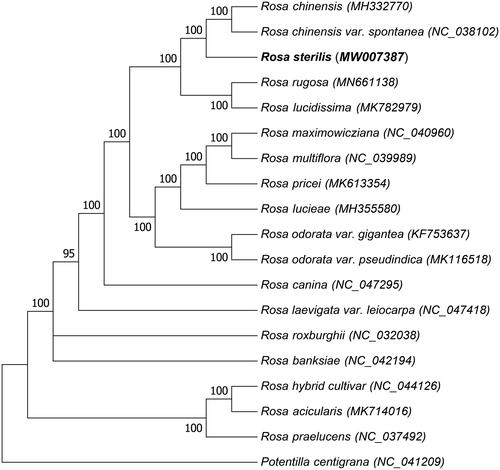
The chloroplast genomes of 18 species and varieties in the Rosaceae and R. sterilis were used to validate the phylogenetic position. The complete chloroplast genomes were aligned by MUSCLE version 3.8.31 (http://www.drive5.com/muscle/) method (Edgar Citation2004). The tree was constructed using the maximum likelihood with MEGA version 7 software, which the bootstrap value was calculated using 1000 replicates (). The phylogenetic tree results showed that R. sterilis is clustered and closest related to Rosa chinensis (MH332770) or Rosa chinensis var. spontanea (NC_038102). The complete chloroplast of R. sterilis can be essential for genomic studies and genetic engineering studies of the family Rosaceae.
Disclosure statement
No conflict of interest was declared by the author(s).
Data availability statement
The complete chloroplast genome sequences of Rosa sterilis together with corresponding annotations are available under the accession number MW007387 (http://www.ncbi.nlm.nih.gov/biosample/16561237). The chloroplast genome raw sequencing reads obtained by Illumina NovaSeq6000 sequencing are available at NCBI Sequence Read Archive (SRA) under the accession number SRR12967098. The Bioproject accession number is PRJNA672238 (http://www.ncbi.nlm.nih.gov/bioproject/672238) and the Biosample accession number is SAMN16561237 (http://www.ncbi.nlm.nih.gov/biosample/16561237).
Additional information
Funding
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- He SL, Yang Y, Li ZW, Wang XJ, Guo YB, Wu HZ. 2020. Comparative analysis of four Zantedeschia chloroplast genomes: expansion and contraction of the IR region, phylogenetic analyses and SSR genetic diversity assessment. PeerJ. 8: e9132.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Lapidus A, Antipov D, Bankevich A, Gurevich A, Korobeynikov A, Nurk S, Prjibelski A, Safonova Y, Vasilinetc I, Pevzner PA. 2014. New Frontiers of Genome Assembly with SPAdes 3.0. (poster). Algorithmic Biology Laboratory, St. Petersburg Academic University, St. Petersburg, Russia. http://bioinf.spbau.ru/spades.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715–721.
- Liu MH, Zhang Q, Zhang YH, Lu XY, Fu WM, He JY. 2016. Chemical analysis of dietary constituents in Rosa roxburghii and Rosa sterilis fruits. Molecules. 21(9):1204–1224.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.
- Luo XL, Dan HL, Li N, Li YH, Zhang YJ, Zhao P. 2017. A new catechin derivative from the fruits of Rosa sterilis S. D. Shi. Nat Prod Res. 31(19):2239–2244.