Abstract
In this study, we sequenced and analyzed the complete mitochondrial genome of Alectryonella plicatula (Gmelin 1791), the newly determined mitochondrial genome is 18225 bp in length, it is a circular molecule and consists of 12 protein-coding genes (atp8 is absent), 24 transfer RNA (with two copies of trnP and trnQ), and 2 ribosomal RNA genes (splitting of the rrnL gene and duplication of the rrnS gene were identified). Phylogenetic analysis based on 12 protein coding genes showed that Alectryonella plicatula is closely related to Crassostrea gigas.
Alectryonella plicatula is a native species that distributed along the coast of China, commonly known as ‘grass oyster’, ‘zhe oyster’. It mainly inhabiting intertidal hard grounds can tolerate a wide range of salinity and temperature (Wang Citation1988). The shell of A. plicatula is relatively thin and often smaller than other cupped oyster Crassostrea species (Yu et al. Citation2008). To date, studies use molecular tools to understand the taxonomic status of A. plicatula and discriminate this species from others have only used partial genomes ( Liu and Dai 1998; Yu et al. Citation2008, Wang et al. Citation2008). The complete mitochondrial genome of A. plicatula is thus of great importance to shed some light on the diversity and phylogenetic relationship of Ostreidae.
In this study, the complete mitochondrial genome of A. plicatula was determined by next-generation sequencing (NGS) platform. Specimen was collected from Shicheng Island, Dalian (122° 98′ 38E, 39° 56′ 69 N) and stored at Liaoning Ocean and Fisheries Science Research Institute, Dalian, China (voucher no. CP-202005). After collection, specimens were immediately preserved in 95% ethanol until DNA extraction. Total genomic DNA was isolated using the DNeasy tissue kit (Qiagen) according to the manufacturer’s instructions. Total genomic DNA was sequenced on an Illumina HiSeq sequencer using a PE150 protocol. The raw reads were filtered using Trimmomatics (Bolger et al. Citation2014), and were assembled using NOVOPlasty (Dierckxsens et al. Citation2017).
The complete mitochondrial genome of A. plicatula (Genbank assession number MW143047) is 18225 bp in length, according to the annotation results from MITOS (Bernt et al. Citation2013) and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/), it contains 12 protein-coding genes (as in most bivalves, the atp8 gene is missing), 24 tRNA genes, 2 rRNA genes, and the splitting of the rrnL gene and duplication of the rrnS gene were identified. Typical metazoan mitogenome have a set of 22 tRNA genes, including two copies of trnL and two of trnS (Podsiadlowski et al. Citation2008), but the number of tRNA genes in oyster mitogenomes varies (Wu et al. Citation2010). The mitogenome of A. plicatula also did not follow that rule, which include extra copies of trnM and trnQ. The nucleotide composition of this mitogenome is A 27.6%, C 14.6%, G 22.0%, T 35.7%, with A + T content (63.3%) higher than G + C content (36.7%), the nucleotide compositions and AT contents of the entire mitogenome is similar to the pattern of other oyster mitogenomes (Wu et al. Citation2010).
The phylogenetic relationships of A. plicatula to Crassostrea species were reconstructed based on the 12 protein coding genes using maximum-likelihood analysis, three members from Ostrea (Bivalvia: Ostreidae) were selected to root the tree (). A. plicatula was recovered as the sister group of C. gigas with the maximal support, thus validated the close relationship between the two species. This clade is then group together with C. angulate, and C. sikamea is the sister to the clade containing C. angulate and C. gigas + A. plicatula.
Figure 1. The maximum-likelihood (ML) phylogenetic relationships of Crassostrea based on the 12 protein coding genes using IQ-TREE v1.6.1(Nguyen et al. Citation2015). The ML searches were run using a combination of rapid hill-climbing approaches and the stochastic perturbation method with 1,000 ultrafast bootstraps.
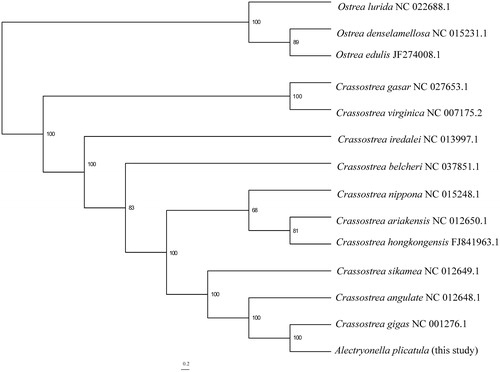
The complete mitogenome of A. plicatula presents here provide valuable resources for accurate molecular barcoding and identification of specimens of this native species. In addition, it will also be useful for investigating the phylogeny and population genetics of Asian oyster species.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW143047, or available from the corresponding author. The raw sequence data were deposited in SRA, accession number SRR13449415.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Podsiadlowski L, Braband A, Mayer G. 2008. The complete mitochondrial genome of the onychophoran Epiperipatus biolleyi reveals a unique transfer RNA set and provides further support for the ecdysozoa hypothesis. Mol Biol Evol. 25(1):42–51.
- Liu BQ, Dai, JXXun DJ. 1998. Studies on genetic diversity in oysters, Crassostrea. Shuichan Xuebao. 22(003):193–198.
- Wang R. 1988. Coloured illustrations of aquatic mollusks in China. Vol. 255. Zhejiang Publishing House of Science and Technology, Hangzhou. (in Chinese)
- Wang H, Zhang G, Liu X, Guo X. 2008. Classification of common oysters from North China. J. Shellfish Res. 27(3):495–503.
- Wu X, Xu X, Yu Z, Wei Z, Xia J. 2010. Comparison of seven Crassostrea mitogenomes and phylogenetic analyses. Mol Phylogenet Evol. 57(1):448–454.
- Yu H, Li Q, Yu R. 2008. Microsatellite variation in the oyster Alectryonella plicatula along the coast of China. J Ocean Univ China. 7(4):432–438.
