Abstract
The white seabream Diplodus sargus (Linnaeus, 1758) is a species of interest for commercial fisheries throughout its range of distribution and it is also reared using aquaculture techniques. Herein, we present the first complete sequence and annotation of the mitochondrial genome of this species. The D. sargus mitogenome is 16,515 base pairs in length and contains 13 protein-coding genes, 2 rRNA, 22 tRNA, and 2 non-coding regions (D-loop and L-origin). The overall nucleotide composition is: 27.3% A, 28.9% C, 26.8% T, and 17.0% G. Maximum likelihood analyses placed D. sargus as a sister species of Diplodus puntazzo. This study provides valuable information for further studying identification methods and evolutionary relationships of Sparidae species.
The white seabream Diplodus sargus (Linnaeus, 1758) is a coastal and gregarious demersal species classified in the family Sparidae Rafinesque, 1818 and is distributed in the Mediterranean Sea and the Black Sea (Fricke et al. Citation2016). Diplodus sargus are usually ∼22 cm in total length (Bauchot Citation1987) and the maximum recorded weight was 1.9 Kg (IGFA Citation2001). These features make this species an appreciable target for fishery activities and also for aquaculture purposes in the Mediterranean basin (Karakatsouli et al. Citation2007). Herein, we present the first characterization of the complete mitochondrial genome sequence of D. sargus (GenBank Accession: MW559786).
Herein, a specimen of white seabream was caught with trammel nets (5–15 m depth) off Posillipo (Naples, Tyrrhenian Sea, Mediterranean Sea, ∼40°48′38′′N, 14°12′28′′E) on 9 February 2020. It was identified on the basis of species-specific diagnostic characters and subsequently deposited in the Darwin Dohrn Museum of the Stazione Zoologica Anton Dohrn of Naples (http://www.szn.it/index.php/en/museum-archives-library/darwin-dohrn-museum, curator Andrea Travaglini, [email protected]) with the code number SZN-OST-0002. Mitochondrial DNA was extracted from dorsal fin tissue following the method described in Mascolo, Ceruso, Sordino, et al. (Citation2019). The assembled mitogenome was obtained by high-throughput sequencing of enriched mitochondrial DNA with Illumina NextSeq 550 System (Illumina, San Diego, CA). Bioinformatics analyses were performed at the BIOINforMA service of the Stazione Zoologica Anton Dohrn (Naples, Italy). Contigs and scaffolds were assembled using the MetaSPAdes version 3.12 tool (Nurk et al. Citation2017). Scaffold assignation to mitochondrial genome was performed by BLAST-based searches against known fish mitochondrial genomes using USEARCH version 11 (Edgar Citation2010). Prediction and annotation of genes were performed using the Prokka version 3.2.1 tool (Seemann Citation2014).
The D. sargus mitogenome is 16,515 base pairs (bp) long, and contains 13 protein-coding genes, 2 ribosomal RNA (12S and 16S), 22 transfer RNA (tRNA), and 2 non-coding regions (D-loop and L-origin) in agreement with Fietz et al. (Citation2020). The mitochondrial structure and gene organization are very similar to those of the sister species Diplodus puntazzo (Walbaum, 1792) (see Ceruso et al. Citation2020). All mitochondrial genes are encoded on the heavy strand, with the exception of the NADH dehydrogenase subunit 6 gene (ND6) and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser [UCN], and Glu, Pro) which are encoded on the light strand. The overall base composition is 27.3% A, 28.9% C, 26.8% T, and 17.0% G, similar to that observed in other species of the same family (Ceruso, Mascolo, Lowe, et al. Citation2018, Ceruso, Mascolo, Palma, et al. Citation2018; Mascolo et al. Citation2018a, Citation2018b, Mascolo, Ceruso, Chirollo, et al. Citation2019; Ceruso et al. Citation2020). All protein-coding genes initiate with an ATG start codon except COI, which starts with GTG. We identified five types of stop codons, namely TAA (ND1, ND4L, and ND5), AGG (COI), T (CYTB, ND3, COII, and ND4), TAG (ATP8 and ND6), and TA (ATP6, COIII, and ND2). The 12S and 16S rRNA genes were located between the tRNAPhe (GAA) and tRNALeu (TAA) genes and were separated by the tRNAVal gene as reported in other vertebrates (Li et al. Citation2016). The 22 tRNA genes vary from 67 (tRNACys) to 74 bp (tRNALts) in length. The control region (846 bp) is located between tRNAPro (TGG) and tRNAPhe (GAA). The non-coding region (L-strand origin of replication) is 32 bp long and is located between tRNAAsn (GTT) and tRNACys (GCA).
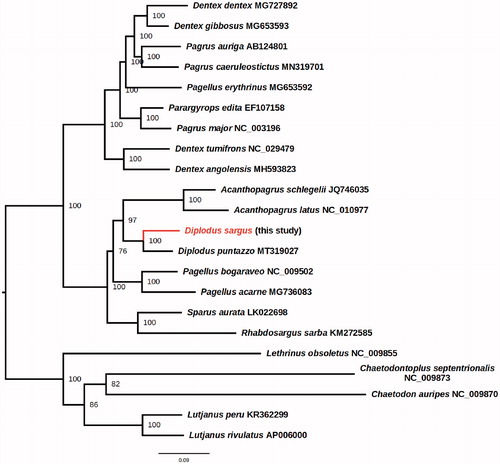
The phylogenetic position of D. sargus within the Sparidae family was inferred with maximum likelihood approach using IQ-TREE 2 (Minh et al. Citation2020) with the ultrafast bootstrap implementation with 1000 replicates (Hoang et al. Citation2018), using the TIM2 + F + I+G4 nucleotide model chosen according to the Bayesian information Criterion by fast model selection (Kalyaanamoorthy et al. Citation2017). These results place D. sargus as a sister species of D. puntazzo in a well-supported clade with Acanthopagrus, confirming the generic placement inferred by Ceruso et al. (Citation2020) (). In the phylogeny herein presented, Diplodus and Acanthopagrus are sister genera. This is in contrast with a recently Cyt-b based barcode phylogeny of the Sparidae family (Ibrahim et al. Citation2020), but in agreement with the COI-based barcode phylogeny of Rabaoui et al. (Citation2019), highlighting the importance of mitogenome-based phylogenetic studies for the understanding of Sparidae evolutionary relationships. This study provides new molecular data for studying this marine fish species of commercial and nutritional importance.
Figure 1. Phylogenetic position of D. sargus within the family Sparidae. Accession number of the Sparidae mitochondrial genome sequences herein used are: Acanthopagrus latus NC_010977, Acanthopagrus schlegelii JQ746035, Dentex angolensis MH593823, Dentex dentex MG727892, Dentex gibbosus MG653593, Dentex tumifrons NC_029479, Pagellus acarne MG736083, Pagellus bogaraveo NC_009502, Pagellus erythrinus MG653592, Pagrus auriga AB124801, Pagrus caeruleostictus MN319701, Pagrus major NC_003196, Parargyrops edita EF107158, Rhabdosargus sarba KM272585, Sparus aurata LK022698, Diplodus puntazzo MT319027. Five outgroup species (Chaetodon auripes NC_009870, Chaetodontoplus septentrionalis NC_009873, Lethrinus obsoletus NC_009855, Lutjanus peru KR362299, and Lutjanus rivulatus AP006000) were selected. Maximum likelihood method was used with an automatic bootstrapping cutoff of 0.01.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW559786.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA708158, SRX10291858, and SAMN18220291, respectively.
References
- Bauchot ML. 1987. Poissons osseux. In Fischer W, Bauchot ML, Schneider M, editors. Fiches FAO d’identification pour les besoins de la pêche. (rev. 1). Méditerranée et mer Noire. Zone de pêche 37. Vol. II. Rome: Commission des Communautés Européennes and FAO; p. 891–1421.
- Ceruso M, Mascolo C, Lowe EK, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common Pandora Pagellus erythrinus (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(2):624–625.
- Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common dentex, Dentex dentex (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(1):391–392.
- Ceruso M, Venuti I, Osca D, Caputi L, Anastasio A, Crocetta F, Sordino P, Pepe T. 2020. The complete mitochondrial genome of the sharpsnout seabream Diplodus puntazzo (Perciformes: Sparidae). Mitochondrial DNA B Resour. 5(3):2379–2381.
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26(19):2460–2461.
- Fietz K, Trofimenko E, Guerin PE, Arnal V, Torres-Oliv M, Lobréaux S, Pérez-Ruzafa A, Manel S, Puebla O. 2020. New genomic resources for three exploited Mediterranean fishes. Genomics. 112(6):4297–4303.
- Fricke R, Golani D, Appelbaum-Golani B. 2016. Diplodus levantinus (Teleostei: Spari-dae), a new species of sea bream from the southeastern Mediterranean Sea of Israel, with a checklist and a key to the species of the Diplodus sargus species group. Sci Mar. 80(3):305–320.
- Hoang DT, Chernomor O, Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Ibrahim N, M. Abbas E, El-Seedy A, Soliman T, S. Ali F. 2020. DNA barcoding and comparative genetic diversification among species of family Sparidae in the coastal waters of Egypt. Egypt J Aquatic Biol Fish. 24(3):333–349.
- IGFA 2001. Database of IGFA angling records until 2001. Fort Lauderdale (FL): IGFA. http://www.igfa.org/.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von A, Jermiin LS. 2017. Model finder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Karakatsouli N, Papoutsoglou SE, Manolessos G. 2007. Combined effects of rearing density and tank colour on the growth and welfare of juvenile white sea bream Diplodus sargus L. in a recirculating water system. Aquacult Res. 38(11):1152–1160.
- Li J, Yang H, Xie Z, Yang X, Xiao L, Wang X, Li S, Chen M, Zhao H, Zhang Y. 2016. The complete mitochondrial genome of the Rhabdosargus sarba (Perciformes: Sparidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):1606–1607.
- Mascolo C, Ceruso M, Chirollo C, Palma G, Anastasio A, Sordino P, Pepe T. 2019. The complete mitochondrial genome of the Angolan dentex Dentex angolensis (Perciformes: Sparidae). Mitochondrial DNA B. 4(1):1245–1246.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018a. The complete mitochondrial genome of the axillary seabream, Pagellus acarne (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(1):434–435.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018b. The complete mitochondrial genome of the Pink dentex Dentex gibbosus (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(2):525–526.
- Mascolo C, Ceruso M, Sordino P, Palma G, Anastasio A, Pepe T. 2019. Comparison of mitochondrial DNA enrichment and sequencing methods from fish tissue. Food Chem. 294:333–338.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Haeseler MD, Von A, Lanfear R. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Rabaoui L, Yacoubi L, Sanna D, Casu M, Scarpa F, Lin YJ, Shen KN, Clardy TR, Arculeo M, Qurban MA. 2019. DNA barcoding of marine fishes from Saudi Arabian waters of the Gulf. J Fish Biol. 95(5):1286–1297.
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30(14):2068–2069.
