Abstract
In the present study, we obtained and annotated the complete mitochondrial genome (mitogenome) of Traulia minuta. The length of the whole mitogenome was 15,636 bp and the AT content of the complete mitogenome was 74.5%. All protein-coding genes (PCGs) started with typical ATN codon and ended with complete TAA/TAG codons except Nad5, which ended with incomplete T codon. The phylogenetic tree indicated that T. minuta was clustered together with T. szetschuanensis.
Traulia minuta Huang & Xia, 1985 is a grasshopper belonging to the Traulia Stål, 1873, Catantopinae, Acridoidae, Acridoidea, Orthoptera (Cigliano et al. Citation2020). The Traulia Stål, 1873, contained 54 species, distributed in Asia (Cigliano et al. Citation2020). Here, we obtained and annotated the whole complete mitochondrial genome (mitogenome) of T. minuta, with the accession number, MF113247. The sample of T. minuta was collected from Lai yang river (Si Mao, China) (101°19 N, 22°57 E) in 2007, and was deposited in Molecular and Evolutionary Lab in Shaanxi Normal University in China and the voucher number of the specimen was 20070728M1. The T. minuta mitogenome sequences were assembled with the Staden Package 1.7 (Staden et al. Citation2000). Geneious Prime (Kearse et al. Citation2012) and MITO (Bernt et al. Citation2013) were used for protein-coding genes (PCGs) annotation. tRNAscan-SE 2.0 (Lowe and Chan Citation2016), MITO (Bernt et al. Citation2013), and Geneious Prime (Kearse et al. Citation2012) were used for tRNA genes annotation. Two rRNAs and the non-coding region were confirmed by similarity blast with related species using Geneious Prime (Kearse et al. Citation2012).
The mitochondrial genome of T. minuta was a closed circular double-stranded structure with the length of 15,636 bp. It contained 13 PCGs (COX1-3, ND1-6, ND4L, Cytb, ATP6, and ATP8), 22 tRNAs, and two rRNAs (rrnS and rrnL) and a non-coding region. Among them, 14 genes (including four PCGs, eight tRNAs, and two rRNAs) are encoded in the N-strand, and the remaining 23 genes are encoded in the J-strand. AT content of the whole sequences was 74.5% (A 42.4%, T 32.1%, C 14.7%, and G 10.8%). The mitogenome had a compact structure with 11 overlaps, ranging from 1 to 8 bp in length. Two rRNA genes, rrnS and rrnL were 796 bp and 1317 bp in size, respectively. The non-coding region was 784 bp in length and the AT content was as high as 82.9%.
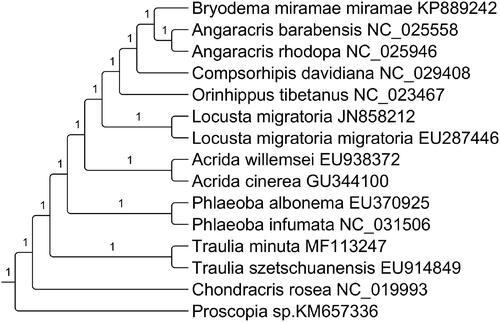
Thirteen PCGs were concatenated by SequenceMatrix v1.7.8 (Gaurav et al. Citation2011). We construct phylogenetic relationship based on 13 PCGs with related 15 species using the maximum-likelihood (ML) method by RAxML (Stamatakis Citation2006) with 1000 bootstrap replicated and GTR + G + R substitution model (). The phylogenetic analysis showed that T. minuta was clustered together with T. szetschuanensis.
Disclosure statement
The authors report no conflicts of interest.
Data availability statement
The data that support the findings of the present study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MF113247, with the accession number, MF113247.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cigliano MM, Braun H, Eades DC, Otte D. 2020. Orthoptera species file. Version 5.0/5.0. [11.X.2019]. http://Orthoptera.SpeciesFile.
- Gaurav V, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Staden R, Beal KF, Bonfield JK. 2000. The Staden package, 1998. Methods Mol Biol. 132:115–130.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.