Abstract
Agelasta perplexa Pascoe is a mulberry borer that threatens the health of the plant. This study revealed the length of the complete mitochondrial genome of A. perplexa which consists of 15,552 bp length with 39.8% A, 12.8% C, 8.4% G, and 39.1% T, respectively. The GC content of whole mitochondrial genome is 21.1%. The complete mitochondrial genome encodes 12 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and one AT-rich region. This study can facilitate further research about genetic evolution as well as prevention and control strategy of A. perplexa.
Agelasta perplexa Pascoe (Coleoptera: Cerambycidae) is mainly distributed in Southeast China. The dark body is 11–18 mm long with intensive reddish-brown and gray-white spots. A. perplexa is predominant pest of Morus alba L., causing extensive damage to the plant (Yamasako and Ohbayashi Citation2012), so it is particularly important to control it. However, species information of A. perplexa is not enough. In order to better know and reveal the genetic and evolutionary information of A. perplexa, so as to better carry out the prevention and treatment work, the complete mitochondrial genome of A. perplexa was determined in this study.
There were five adults of A. perplexa collected from Lianjiang, Fujian province, China (26.32836 N, 119.80018 E) by the traps with sex pheromone. The specimens were preserved at −80 °C in the Key Laboratory of Integrated Pest Management in Ecological Forests, Fujian Agriculture and Forestry University (Songqing Wu, [email protected]) under the voucher number TN-202010. To construct the sequencing library and obtain sequence information from mitochondrial DNA, the TruSeq DNA Sample Preparation Kit (Vanzyme, Nanjing, China) was utilized to extract the total DNA and the samples were purified by the QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). Then, the DNA library was contrusted using transposase method. Agencourt Spriselect was used to purify the library and perform size selection, and the library with a fragment peak value of 300 bp was selected for sequencing. Furthermore, the mitochondrial genome was 2 × 150 paired-end sequenced by the Illumina Hiseq 2500 (Illumina, San Diego, CA) at Genesky Biotechnologies Inc. (Shanghai, China). After quality control and filtration, the total 58,116,830 clean reads were obtained from 60,728,446 raw reads. Then, MitoZ and metaSPAdes were applied to assemble the clean reads (Nurk et al. Citation2017). GeSeq was used to annotate the mitogenome of samples followed by individually correction in Geneious using a reference genome (GenBank accession no. KY292221) (Masters et al. Citation2015; Michael et al. Citation2017).
The results of assembly and annotation indicated that length of the complete mitochondrial genome of A. perplexa is 15,552 bp. The GC content of whole mitochondrial genome is 21.1%. The complete mitochondrial genome encodes 12 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and a 905 bp AT-rich region. Twelve PCGs are 10,710 bp in total, encoding 3570 amino acids. Six PCGs (COII, COIII, ND3, ND4, ND4L, CYTB) start with a typical ATN codon, while the other PCGs codons are unusual; nine PCGs (ND2, COI, COII, ATP6, COIII, ND5, ND4, ND4L, ND6) stop with codon TAA, and three PCGs (ND3, CYTB, ND1) stop with codon TAG. The rrnS and rrnL genes are 781 bp and 1275 bp in length, respectively.
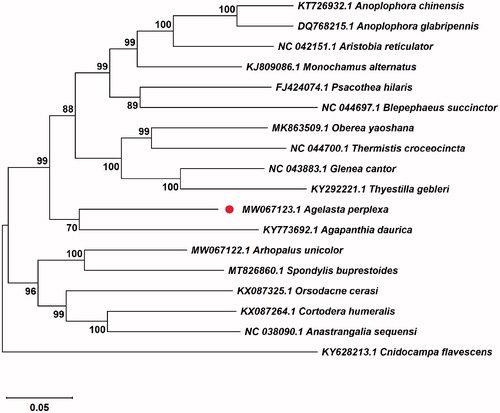
In order to gain insight into the phylogenetic relationships of A. perplexa, MAFFT 7.0 (Kazutaka and Standley Citation2013) was used for sequence alignment according to the complete mitochondrial genome sequence of A. perplexa. Then using a Lepidoptera species Cnidocampa flavescens Walker (GenBank accession no. KY628213) as an out-group, a maximum-likelihood phylogenetic tree with 1000 bootstraps was constructed using MEGA 7.0 to reconstruct the phylogenetic position of A. perplexa with 16 related Coleoptera species (Kumar et al. Citation2016). The resultant ML trees showed that the A. perplexa was clustered together with Agapanthia daurica Ganglbauer, and constituted a monophyletic group with 10 species belonging to the subfamily Lamiinae. The monophyletic Lamiinae species was assigned to the sister group to the clade of Orsodacnidae and Cerambycidae that consists of Orsodacne, Aseminae, Lepturinae, and Spondylinae in this study (). The results consolidate the existing knowledge of A. perplexa. The availability of the complete mitochondrial genome of A. perplexa allows further research about the genetic evolution and control tactics of A. perplexa can be facilitated.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/MW067123, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA681753, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN16967690, and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR13172076.
Additional information
Funding
References
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):msw054.
- Masters BC, Fan V, Ross HA. 2015. Species delimitation—a geneious plugin for the exploration of species boundaries. Mol Ecol Resour. 11(1):154–157.
- Michael T, Pascal L, Tommaso P, Ulbricht-Jones ES, Axel F, Ralph B, Greiner S. 2017. Geseq– versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. Metaspades: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Yamasako J, Ohbayashi N. 2012. Taxonomic position of the oriental species of Mesosa (Mesosa) (Coleoptera, Cerambycidae, Lamiinae, Mesosini). Psyche. 2012:1–15.