Abstract
Ziziphus attopensis has high medicinal value of tonic, carminative, appetizer, and muscle analgesic. To provide genetic information for further research and conservation, the complete chloroplast genome sequence of Z. attopensis was determined in this study. The genome was 161,342 bp in length, consisting of a large single copy region (89,035 bp), a small single copy region (19,421 bp), and a pair of inverted repeat regions (26,443 bp). In total, 129 genes were identified in Z. attopensis chloroplast genome, including 85 protein coding genes, eight rRNA, and 36 tRNA genes. Maximum-likelihood tree inferred from 58-gene sequences of 20 taxa supported that Rhamnaceae is sister to Elaeagnaceae and Z. attopensis is more closely related to Ziziphus jujuba.
Ziziphus attopensis Pierre is a scandent shrub belonging to the family of Rhamnaceae (Hauenschild et al. Citation2016), distributed in sparse forests, thickets, up to 1500 m in Liaos and Thailand. The decoction from the bark and wood of Z. attopensis was suggested to be used as tonic, carminative, appetizer, and muscle analgesic. The water extract from Z. attopensis did not contain acute or chronic toxicities (Sireeratawong et al. Citation2012). In this study, the complete chloroplast genome of Z. attopensis was determined and analyzed.
Fresh leaves of Z. attopensis were collected from Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (N21°55′53″, E101°15′46″) and stored in silica gel for later use. The voucher specimen (MGZ-01) was deposited at Shaanxi Xueqian Normal University. Total DNA was extracted from 2 g of leaves using Qiagen DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Sequencing was conducted on Illumina HiSeq™ 2500 platform (Illumina Inc., San Diego, CA). After removing adapter and low-quality reads, contigs were de novo assembled using velvet (V1.2.07) (Zerbino and Birney Citation2008). Complete chloroplast genome of Z. attopensis was annotated by PGA (Qu et al. Citation2019) and submitted to GenBank using Bankit (The accession number is MW201670).
Assembled chloroplast genome of Z. attopensis was 161,342 bp in size, which is shorter than Ziziphus jujuba with 161,466 bp and Z. jujuba cv. Dongzao with 161,493 bp (Gao et al. Citation2017; Ma et al. Citation2017). It has a typical quadripartite circular structure including a larger single copy region with 89,035 bp, a small single copy region with 19,421 bp separated by a pair of inverted repeat sequences with 26,443 bp. A total of 129 genes were identified, including 85 protein-coding genes, eight rRNA genes, and 36 tRNA genes. Among which, five protein-coding, four rRNA, and six tRNA genes were duplicated in IR regions. Fifteen genes contain one intron (trnK-UUU, rps16, rpoC1, atpF, trnG-UCC, trnL-UAA, trnV-UAC, petD, petB, rpl16, rpl2, ndhB, trnA-UGC, ndhA, trnI-GAU), and three genes contain two introns (ycf3, clpP, rps12). InfA gene has been lost in Z. attopensis cp genome. Overall GC content of the cp genome is 36.81%, the LSC, SSC, and IR are 34.67%, 30.87%, and 42.60%, respectively.
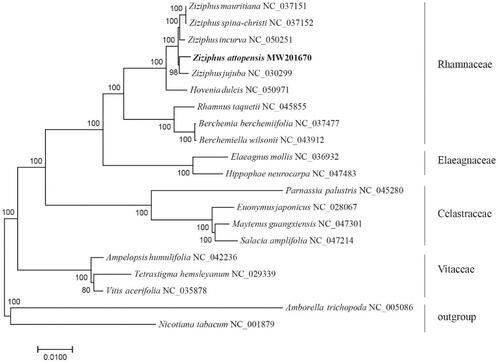
To investigate the phylogenetic position of Z. attopensis, maximum-likelihood implemented in MEGA (Kumar et al. Citation2018) was used to construct the phylogenetic tree. As the chloroplast genome structure may be rearranged among different species, analysis was performed using 58 protein-coding genes that were present in 18 species from Rhamnaceae, Vitaceae, Celastraceae, Elaeagnaceae, and two outgroups Nicotiana tabacum and Amborella trichopoda. Result revealed that Z. attopensis clustered with Z. jujuba; five species from Ziziphus genus consisting of a monophyletic group, and then clustered with Hovenia dulcis. Rhamnaceae was resolved as sister to Elaeagnaceae (). This study provides basic data for the research of genetic diversity and phylogeography of Z. attopensis population and supplies evidence for the phylogeny of Rhamnaceae in aspect of molecular data.
Acknowledgements
We thank Houcheng Xi from Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences for collecting samples; Xin Zhou from Shaanxi Xueqian Normal University for data analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly sequenced in this study are available in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov) under accession number MW201670. The raw sequence data were deposited successfully with registered number of SRR13442540.
Additional information
Funding
References
- Gao CM, Gao YC, Liu XH. 2017. The complete genome of Ziziphus jujuba cv. dongzao, an economic crop in Yellow River Delta of China. Mitochondrial DNA Part B. 2(2):692–693.
- Hauenschild F, Matuszak S, Muellner-Riehl AN, Favre A. 2016. Phylogenetic relationships within the cosmopolitan buckthorn family (Rhamnaceae) support the resurrection of Sarcomphalus and the description of Pseudoziziphus gen. nov. Taxon. 65(1):47–64.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Ma QY, Li SX, Bi CW, Hao ZD, Sun CR, Ye N. 2017. Complete chloroplast genome sequence of a major economic species, Ziziphus jujuba (Rhamnaceae). Curr Genet. 63(1):117–129.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Sireeratawong S, Vannasiri S, Nanna U, Singhalak T, Jaijoy K. 2012. Evaluation of acute and chronic toxicities of the water extract from Ziziphus attopensis Pierre. ISRN Pharmacol. 2012:789651.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.