Abstract
The mitogenomes of two insular subspecies of Pazala, G. (P.) eurous asakurae and G. (P.) mullah chungianus from Taiwan Island, are reported. Both mitogenomes are circular, 15,228 bp and 15,240 bp in length respectively, and consist of 37 genes, including 13 PCGs, 22 tRNAs, and two rRNAs. The Bayesian phylogenetic tree containing the focal taxa and 31 other Papilioninae members clustered them with G. (P.) mullah (Alphéraky, 1897) and then G. (P.) parus (Nicéville, 1900) inside tribe Leptocircini, which agrees with their taxonomic positions. The findings of this study would benefit future understanding of phylogeography and conservation of subgenus Pazala.
Taiwan Island is the easternmost point in the distribution range of the swordtail butterflies of subgenus Pazala Moore, 1888 (Lepidoptera: Papilionidae), represented by two endemic subspecies, Graphium (Pazala) eurous asakurae (Matsumura, 1908) and G. (P.) mullah chungianus (Murayama, 1961) (Racheli and Cotton Citation2009; Hsu et al. Citation2018). Although the endemicity of some Pazala taxa has been partly discussed in recent taxonomic works (Hu et al. Citation2018, Citation2019; Zhang et al. Citation2020), the phylogeographic history of this intriguing Sino-Himalayan swallowtail group is still poorly understood. The mitogenome contains very useful phylogenetic information for evolution, and has become a feasible tool in phylogeography. The complete mitogenomes of the two insular endemic subspecies of Pazala reported herein would facilitate future research related to in-depth butterfly conservation (Wang et al. Citation2020).
The samples used in this study were collected from Taiwan Island. G. (P.) eurous asakurae came from Taroko, Hualien (24.202133°N, 121.454329°E), and G. (P.) mullah chungianus was collected at Fushan, Yilan (24.749637°N, 121.639251°E). The specimens were deposited in the zoological museum (insect collection) of Yunnan University, Kunming, China (specimen numbers: YNU-LEP-PAP-2019028 and YNU-LEP-PAP-2019102, contact person: Shao-Ji Hu). Genomic DNA was extracted from the thoracic muscle tissue of the adult using phenol-chloroform and isopropanol protocol (Hu et al. Citation2013) and sequenced on an ABI 3730xl automatic sequencer (Applied Biosystems, CA, USA). Resultant gene fragments were assembled using DNAStar (https://www.dnastar.com/) with the previously reported mitogenomes of G. (P.) mullah (Alphéraky, 1897) (KJ472924) (Chen et al. Citation2014) and G. (P.) parus (Nicéville, 1900) (MT198821) (Duan et al. Citation2020) as references. Protein-coding genes (PCGs), transfer RNA genes (tRNAs) and ribosomal RNA genes (rRNAs) were predicted using the web based MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and the Alignments | CDS feature under BLASTn of NCBI (https://blast.ncbi.nlm.nih.gov/).
The complete mitogenomes of G. (P.) eurous asakurae (MW549198) and G. (P.) mullah chungianus (MW549197) are circular, 15,228 bp and 15,240 bp in length respectively. The base composition of G. (P.) eurous asakurae is 39.81% for A, 40.58% for T, 7.85% for G, and 11.76% for C; while that of G. (P.) mullah chungianus is 40.11% for A, 40.85% for T, 7.60% for G, and 11.43% for C. Both mitogenomes contain 37 genes, including 13 PCGs, 22 tRNAs, and two rRNAs, plus a non-coding control region. The plus (+) strands of both mitogenomes encode nine PCGs (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad6, and cob), while the minus (–) strands encode four PCGs (nad5, nad4, nad4l, and nad1). The gene arrangement and character of both genomes agree with recently reported ditrysian Lepidoptera mitogenomes (Cao et al. Citation2012; Wang et al. Citation2019; Chen et al. Citation2020).
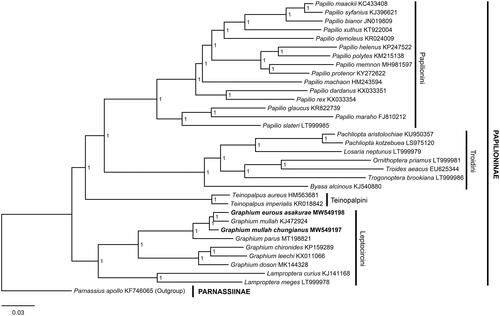
To validate the two mitogenomes, a Bayesian phylogenetic tree was reconstructed using PhyloSuite 1.2.2 (Zhang et al. Citation2020) using the 37 genes (13 PCGs, 22 tRNAs, and two rRNAs) with the GTR + I + G model for 1,000,000 generations. Thirty-one species of Papilioninae with available mitogenomes were used as ingroups and Parnassius apollo Linnaeus, 1758 (Parnassiinae; KF746065) (Chen et al. Citation2014) was chosen as the outgroup. The result clustered the focal subspecies with G. (P.) mullah, while G. (P.) parus is placed in the basal position of Pazala. All Pazala taxa are related to other Graphium species within Leptocircini, forming a monophyletic clade, supported by the maximal support values ().
Acknowledgments
We thank Yu-Feng Hsu, Jia-Yuan Liang, Yu-Chi Lin, and Jung-Chun Lin (National Taiwan Normal University, Taipei, China) for assistance on the collection of specimens, and Adam M. Cotton (Chiang Mai, Thailand) for improving the earlier drafts of this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accession numbers of all used sequences are listed in .
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cao YQ, Ma C, Chen JY, Yang DR. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 13:276.
- Chen L, Wahlberg N, Liao CQ, Wang CB, Ma FZ, Huang GH. 2020. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics. 112:4435–4441.
- Chen YH, Gan SS, Shao LL, Cheng CH, Hao JS. 2014. The complete mitochondrial genome of the Pazala timur (Lepidoptera: Papilionidae: Papilioninae). Mitochondrial DNA Part A. 27(1):533–534.
- Chen YH, Huang DY, Wang YL, Zhu CD, Hao JS. 2014. The complete mitochondrial genome of the endangered Apollo butterfly, Parnassius apollo (Lepidoptera: Papilionidae) and its comparison to other Papilionidae species. J Asia-Pac Entomol. 17:663–671.
- Duan K, Zhang X, Zhang HH, Hu SJ. 2020. Complete mitochondrial genome of the subalpine swordtail butterfly Graphium (Pazala) parus (Nicéville, 1900) (Lepidoptera: Papilionidae). Mitochondrial DNA Part B. 5:1903–1904.
- Hsu YF, Huang CL, Liang JY. 2018. Butterfly Fauna of Taiwan. Vol. 1. Taipei: Council of Agriculture Executive Yuan, Forest Bureau.
- Hu SJ, Condamine FL, Monastyrskii AL, Cotton AM. 2019. A new species of the Graphium (Pazala) mandarinus group from Central Vietnam (Lepidoptera: Papilionidae). Zootaxa. 4554(1):286–300.
- Hu SJ, Cotton AM, Condamine FL, Duan K, Wang RJ, Hsu YF, Zhang X, Cao J. 2018. Revision of Pazala Moore, 1888: the Graphium (Pazala) mandarinus (Oberthür, 1879) group, with treatments of known taxa and descriptions of new species and new subspecies (Lepidoptera: Papilionidae). Zootaxa. 4441(3):401–446.
- Hu SJ, Ning T, Fu DY, Haack RA, Zhang Z, Chen DD, Ma XY, Ye H. 2013. Dispersal of the Japanese pine sawyer, Monochamus alternatus (Coleoptera: Cerambycidae), in mainland China as inferred from molecular data and associations to indices of human activity. PLoS One. 8(2):e0057568.
- Racheli T, Cotton AM. 2009. Guide to the butterflies of the palearctic region. Papilionidae. Part I. Milano: Omnes Artes.
- Wang WL, Suman DO, Zhang HH, Xu ZB, Ma FZ, Hu SJ. 2020. Butterfly conservation in China: from science to action. Insects. 11:661.
- Wang X, Chen ZM, Gu XS, Wang M, Huang GH, Zwick A. 2019. Phylogenetic relationships among Bombycidae s.l. (Lepidoptera) based on analyses of completemitochondrial genomes. Syst Entomol. 44:490–498.
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20:348–355.
- Zhang HH, Cotton AM, Condamine FL, Wang RJ, Hsu YF, Duan K, Zhang X, Hu SJ. 2020. Revision of Pazala Moore, 1888: The Graphium (Pazala) alebion and G. (P.) tamerlanus groups, with notes on taxonomic and distribution confusions (Lepidoptera: Papilionidae). Zootaxa. 4759:77–97.