Abstract
Here, we report the complete chloroplast genome of Keteleeria evelyniana. The genome is 116,940 bp in size, which is comprised of a large single-copy (LSC) region of 74,075 bp, a small single-copy (SSC) region of 40,425 bp, and two short inverted repeat (IR) regions of 1,220 bp. The overall GC content of the plastome was 38.5%. The new sequence comprised 103 unique genes, including 74 protein-coding genes, 4 rRNA genes, and 25 tRNA genes. Phylogenetic analysis showed that K.evelyniana was close to Keteleeria hainanensis and Keteleeria davidiana.
Keteleeria evelyniana is an evergreen tree species belonging to the Pinaceae family, endemic to China. It is mainly distributed in Yunnan, Guizhou and Sichuan in China, occurring in altitude regions between 700 and 2600 m (He et al. Citation2011). It is not only a landscape tree species in Southwest China, but also an economically important tree species in Yunnan. It has been well demonstrated that K.evelyniana is a good material for furniture, due to its wood drying quickly, and easy to process (Zhang et al. Citation2008). Also, K. evelyniana is considered as useful materials for construction, bridges, wooden molds, ships, etc., due to its strong water resistance, moisture resistance and corrosion resistance. Additionally, this tree species is also considered as traditional Chinese medicine in China, that is associated with high medicinal value (Zhang et al. Citation2014).
The complete chloroplast genomes have been proven to be an effective biological tool for rapid and accurate species recognition as super-barcode (Chen et al. Citation2018; Yang et al. Citation2019). To date, many studies have performed whole chloroplast genome sequencing. However, for such an economically important tree species, the classification and phylogenetic relationships of K. evelyniana remain poorly known. In the present study, we report the complete chloroplast genome of K. evelyniana, thus providing useful information for future studies in genetic background and evolution.
The fresh leaves were collected from Southwest Forestry University Kunming, China. (Yunnan, China; geospatial coordinates: 102°45′41″E, 25°04′00″N; Altitude: 1945 m). The voucher specimens of K. evelyniana were deposited at the herbarium of Southwest Forestry University (Voucher number: SWFU-YS-0170) and DNA samples were stored at the Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China Ministry of Education, Southwest Forestry University, Kunming, China. The total genomic DNA was extracted by using the Magnetic beads plant genomic DNA preps Kit (TSINGKE Biological Technology, Beijing, China). A genomic shotgun library with an insert size of 350 bp was prepared and then this library was sequenced and 150 bp paired-end reads were generated. We assembled the chloroplast genome using GetOrganelle v1.6.2e (Jin et al. Citation2020). Genome annotation was performed with the online annotation tool DOGMA (Wyman et al. Citation2004) and program Geneious R8 (Kearse et al. Citation2012). Finally, the chloroplast DNA sequence with complete annotation information was submitted to GenBank with accession number MW043479.
The complete genome of K. evelyniana is 116,940 bp in size with a typical quadripartite structure, containing a small single-copy (SSC) region of 40,425 bp and a large single-copy (LSC) region of 74,075 bp separated by a pair of inverted repeat (IR) regions of 1220 bp. The total GC content is 38.5%, while the corresponding values of the LSC, IR, and SSC regions are 39.2%, 36.6%, and 37.4%, respectively. There were 103 unique genes, including 74 protein-coding genes, 4 rRNA genes, and 25 tRNA genes. Among them 12 genes (atpF, rpoC1, petB, petD, rpl16, rpl2, trnL-UAA, trnK-UUU, trnV-UAC, trnG-UCC, trnI-GAU, and trnA-UGC) have single introns, while one gene(ycf3) have double introns.
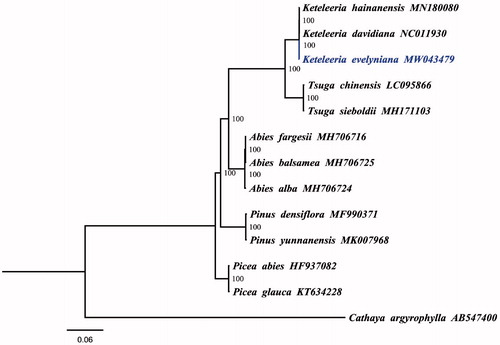
To explore the phylogenetic relationship of K. evelyniana in Pinaceae family, a maximum likelihood tree was constructed based on the complete chloroplast genomes of K. evelyniana and other 12 Pinaceae species. We aligned all chloroplast genomes by using MAFFT v7.307 (Katoh and Standley Citation2013) and analyzed by IQ-TREE 1.5.5 (Nguyen et al. Citation2015) under the TVM + F+R3 model. As illustrated in , K. evelyniana appeared to be closely related to Keteleeria hainanensis and Keteleeria davidiana. The chloroplast genome of K. evelyniana will provide useful genetic information for further study on genetic diversity and conservation of Pinaceae species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number: MW043479. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA687643, SRR13354461, and SAMN17151149 respectively.
Additional information
Funding
References
- Chen XL, Zhou JG, Cui YX, Wang Y, Duan BZ, Yao H. 2018. Identifification of ligularia herbs using the complete chloroplast genome as a super-barcode. Front Pharmacol. 9:695.
- He WJ, Zhao HF, Hong JH, He Y, Guang ZZ, Chang JJ, Hong BC, Yu MZ, Ning HT. 2011. Benzoic acid allopyranosides and lignan glycosides from the twigs of Keteleeria evelyniana. Zeitschr Naturforschung B. 66(7):733–739.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21 : 241. DOI:10.1186/s13059-020-02154-5
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang J, Feng L, Yue M, He YL, Zhao GF, Li ZH. 2019. Species delimitation and interspecific relationships of the endangered herb genus Notopterygium inferred from multilocus variations. Mol. Phylogenet. Evolut. 133:142–151.
- Zhang HB, Yang MX, Tu R, Gao L, Zhao ZW. 2008. Fungal communities in decaying sapwood and heartwood of a conifer Keteleeria evelyniana. Curr Microbiol. 56(4):358–362.
- Zhang XH, Xu F, Yang YM, Zhang M, Zhang Y, Bai H, Wang Q. 2014. Analysis and evaluation of nutritional components and total flavonoids in burgeon of Keteleeria evelyniana. Mast Food Res Dev. 35(20):109–111.