Abstract
The Black Drongo Dicrurus macrocercus is a bird belonging to the group Passeriformes, distributed almost all over the country in China. The conservation status of this species is Least Concern (LC) in IUCN. In this study, the complete mitogenome of D. macrocercus was determined. The mitochondrial DNA is packaged in a compact 17,056 based pair (bp) circular molecule with A + T content of 57.04%. It contains 37 typical mitochondrial genes, including 13 protein-coding genes, 2 rRNAs and 22 tRNAs, and 1 non-coding regions. We reconstructed a phylogenetic tree based on mitogenome sequences of 15 Dicruridae species and one outgroup. In trees, D. macrocercus was clustered as an independent clade with high support value (100). The new mitogenome data would provide useful information for application in conservation genetics and further clarify the phylogenetic evolution of this species.
The Black Drongo Dicrurus macrocercus is a bird with a black body and a blue-green metallic luster, which belongs to the group Passeriformes. There are seven subspecies of Black Drongo all over the world, distributed in South Asia and Southeast Asia, and three subspecies distributed in China, southern Tibet subspecies (D. macrocercus albirictus), Taiwan subspecies (D. macrocercus harterti), and common subspecies (D. macrocercus cathoecus). The conservation status of this species is Least Concern (LC) in IUCN. In China, the species also has been listed as a Least Concern (LC) species by the red list of China’s vertebrates (Jiang et al. Citation2016). This species has an extremely large range, and, hence, does not approach the thresholds for Vulnerable under the range size criterion, the population size has not been quantified, but it is not believed to approach the thresholds for Vulnerable under the population size criterion. The Black Drongo is full migrant, breeding in Afghanistan, Guam and Northern Mariana Islands, and resident in Bangladesh, Bhutan, Cambodia, China, etc. and it is distributed almost all over the country in China, they are living in different types of shrubland, grassland, savanna or artificial, terrestrial inhabitants, and does not normally occur in forest. The Black Drongo is aggressive and fierce in nature, they live in pairs or small groups, and mainly feed on insects, such as beetles, dragonflies, cicadas, and ants, and like to rest on tall trees or wires, fly down to catch the prey rapidly, and then fly to a high place directly (Zhao Citation2001).
Up to now, no complete mitochondrial genome data of D. macrocercus are available in the GenBank. In this study, we sequenced the complete mitochondrial genome of D. macrocercus (GenBank number: BankIt2405244 GZUNZ20201129001 MW307917) and examined its phylogenetic relationship with other Passeriformes species whose mtDNA data are available.
The tissues were collected from an accidental death of individual on 21 November 2019, collected in Longdongbao Airport of Guiyang, Guizhou Province, China, the sample was stored under the number GZUNZ20201129001 at the Research Center for Biodiversity and Nature Conservation of Guizhou University. The extraction was performed using DNA Rapid Extraction Kit (Beijing Aidlab Biotechnologies Co., Ltd, Beijing, China) according to the kit manual. The mitochondrial genomes of D. hottentottus (NC_043948.1) was used to design primers for polymerase chain reaction (PCR) and used as a template for gene annotation.
The complete mtDNA sequences of the Black Drongo were 17,056 bp in length, its overall base composition was: A, 32.19%; C, 28.64%; G, 14.32%, and T, 24.85%. The A + T content was 57.04%, out the range for avian mitogenomes (51.6–55.7%; Haring et al. Citation2001). After quality-proofing of the obtained fragments, the two mitogenomic sequences were assembled manually using DNAstar v7.1 software (Keyser Citation2013). First, raw mitogenomic sequences were imported into MITOS web serves (Bernt et al. Citation2012) to determine the approximate boundaries of genes. Exact positions of protein-coding genes (PCGs) were found by searching for ORFs (employing genetic code 9, echinoderm, and flatworm mitochondrion). All tRNAs were identified using ARWEN (Laslett and Canbck Citation2008), DOGMA (Wyman et al. Citation2004), and MITOS. The precise boundaries of rrnl and rrnS were determined via a comparison with homologs. It has a typical circular mitochondrial genome containing 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs, and 1 non-coding A + T-rich region, which is usually found in birds (Boore Citation1999). The order and orientation are identical with the standard avian gene order (Gibb et al. Citation2007). Of the 13 protein-coding genes, 12 utilize the standard mitochondrial start codon ATG; however, COI use GTG as the initiation codon. TAA is the most frequent stop codon, although COII, COIII, and ND4 end with the single nucleotide T, ND1 and ND5 end with AGA, COI end with AGG, ND6 end with TAG ().
Table 1. Organization of the complete mitochondrial genome of Black Drongo Dicrurus macrocercus.
The 12S rRNA is 984 bp, and the 16S rRNA is 1601 bp in length, which are located between tRNA-Phe(gaa) and tRNA-Leu(taa), and separated by tRNA-Val(tac). All tRNAs possess the classic clover leaf secondary structure, as observed in other bird mitogenomes (Bernt et al. Citation2012). Most of the mitochondrial genes are encoded on heavy strand (H-strand) except for ND6 and eight tRNA genes, which are encoded on light strand (L-strand) ().
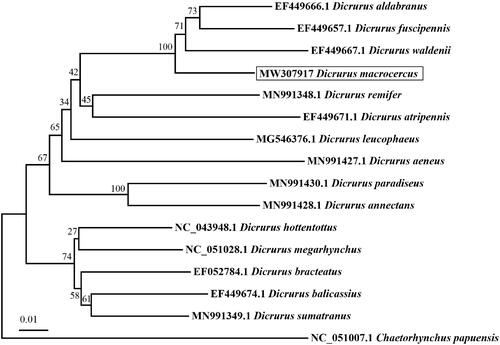
We used MEGA7 to construct the phylogenetic tree using the maximum-likelihood method based on the mitogenome sequences of D. macrocercus and other 15 Dicruridae species and one outgroup by the maximum-likelihood method. In trees, D. macrocercus was clustered as an independent clade with high support value (100) ().
Figure 1. Maximum-likelihood tree based on mitogenome sequences of 15 Dicruridae species and one outgroup. Numbers nodes are bootstrap supports.
This study is the first to report and analyze the complete mitochondrial genome of Black Drongo D. macrocercus. The complete mitogenome of D. macrocercus is in favor of conservation of the non-flagship species, and provides fundamental genetic data for the evolutionary research of Passeriformes.
Acknowledgments
The authors would like to express our gratitude to editors and the anonymous reviewers for their critical comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MW307917; Figure, https://doi.org/10.6084/m9.figshare.13377656.
Additional information
Funding
References
- Bernt M, Braband A, Schierwater B, Stadler PF. 2012. Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol. 69:328–338.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2012. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69:313–319.
- Boore JL. 1999. Survey and summary animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. 2007. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 24:269–280.
- Haring E, Kruckenhauser L, Gamauf A, Riesing MJ, Pinsker W. 2001. The complete sequence of the mitochondrial genome of Buteo buteo (Aves, Accipitridae) indicates an early split in the phylogeny of raptors. Mol Biol Evol. 18:1892–1904.
- Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, Xie F, Cai B, Cao L, Zheng G, et al. 2016. Red list of China’s vertebrates. Biodivers Conserv. 24(5): 500–551.
- Keyser M. 2013. DNASTAR Lasergene: an integrated package for de novo transcriptome and RNA-Seq analysis on a Desktop Computer[C]//International Plant and Animal Genome Conference XXI 2013.
- Laslett D, Canbck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Wyman S, Jansen R, Boore J. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhao ZJ. 2001. A handbook of the birds of China. Jilin, China: Jilin Science and Technology Press. p. 154–155.
