Abstract
Fragaria orientalis Lozinsk. is valuable germplasm material for cross breeding in Fragaria. In this study, we assembled the complete mitochondrial genome of F. orientalis using a combination of Illumina data and Nanopore data. The mitochondrial genome was 275,143 bp in length, including 29 protein-coding genes, 20 tRNA genes, and three rRNA genes, with a total GC content 45.23%. Seven protein-coding genes contained introns, and three were trans-spliced. Phylogenetic analysis indicated that F. orientalis is making a sister clade to the Amygdaloideae species. The complete mitochondrial genome of F. orientalis reported in this study will improve our understanding of Fragaria evolution.
China is among the most diverse centers of wild strawberry resources, with 14 wild species, including 9 diploid species and all 5 tetraploid species (Sun et al. Citation2021). Fragaria orientalis is a wild tetraploid strawberry, that is distributed mainly in Heilongjiang, Jilin and Liaoning Provinces of Northeast China (The CAS Editorial Committee of the Flora of China Citation1985; Lei et al. Citation2017). F. orientalis is potentially valuable as breeding material for its aromatic fruit and resistance to low temperatures (Lei et al. Citation2017). In recent years, breeders have performed interspecific hybridization using F. orientalis to improve cold resistance in Fragaria (Luo et al. Citation2018). In this study, we report the complete mitochondrial genome of F. orientalis for the first time.
Plant materials were harvested from the China National Strawberry Germplasm Repository (Beijing) (39.9°N, 116.2°E). The specimen and DNA were deposited at the Beijing Academy of Forestry and Pomology Sciences (Yong-Shun Gao, [email protected]) under the voucher number DF2020-23. Mitochondrial DNA was extracted from fresh leaves using a plant mitochondrial extraction kit (Balb Co., Ltd, Beijing, China) and then sequenced on Illumina NovaSeq 6000 platform and Nanopore platform, respectively. Approximately 4.8 Gb of raw data were generated with insert-size 450 bp paired-end read lengths on the Illumina sequencing platform. A total of 59 Mb long-reads data comprising 23,812 reads were generated on the Nanopore sequencing platform. The mitochondrial genome was de novo assembled using the Illumina data by GetOrganelle v1.64 (Jin et al. Citation2020), and then the assembled scaffolds were aligned against the Nanopore data. All aligned Nanopore reads were extracted and reassembled with the Illumina data by SPAdes v3.13.1 (Antipov et al. Citation2016). Genome annotation was performed by homologous comparison using BLAST + 2.7.1 (Camacho et al. Citation2008). The tRNA genes were further annotated using tRNAscan-SE v2.07 (Lowe and Eddy Citation1997). The annotated mitochondrial genome has been deposited into GenBank under accession number MW464867. The genome sequences were aligned using MAFFT v7.471 (Katoh et al. Citation2019). Phylogenetic analysis were performed by the maximum likelihood method using RAxML v8.2.12 (Stamatakis Citation2014) based on GTRGAMMA model with 1,000 bootstrap replicates.
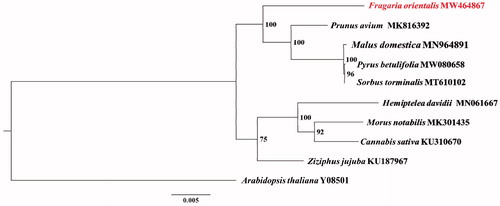
The F. orientalis mitochondrial genome was assembled into a single circular-mapping molecule of 275,143 bp, with a GC content of 45.23%. The mitochondrial genome contained a total of 52 genes, including 29 protein-coding genes, 20 tRNA genes, and three rRNA genes. Among the 29 protein-coding genes, seven genes (nad1, nad2, nad4, nad5, nad7, ccmFc and rps3) contained introns, and three genes (nad1, nad2, nad5) were trans-spliced. Three genes (atp6, nad4L and rps1) used ACG as the start codon, whereas nad1 and mttB used GTG and TTG, respectively. As the stop codon, fifteen genes ended with TAA, seven genes (atp4, ccmB, ccmFn, cob, cox3, nad4 and rps13) with TGA, and seven genes (atp6, ccmC, matR, mttB, nad7, rps3 and sdh4) with TAG. Sequences of 12 common protein-coding genes (atp1, atp9, cox1, cox2, matR, nad2, nad3, nad4, nad4L, nad6, nad7 and nad9) were aligned with homologous genes from 10 species. Phylogenetic analysis showed that F. orientalis is making an isolated clade, which is a sister to the clade making by species from Amygdaloideae (). The complete mitochondrial genome of F. orientalis reported in this study will improve our understanding of Fragaria evolution.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW464867. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA693900, SRR13501403, SRR13501404, and SAMN17487953, respectively.
Additional information
Funding
References
- Antipov D, Korobeynikov A, Mclean JS, Pevzner PA. 2016. HYBRIDSPADES: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 32(7):1009–1015.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2008. BLAST+: architecture and applications. BMC Bioinform. 10:421.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20:1160–1166.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241.
- Lei JJ, Xue L, Guo RX, Dai HP. 2017. The Fragaria species native to China and their geographical distribution. Acta Hortic. 1156(1156):37–46.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Luo GJ, Xue L, Guo RX, Ding Y, Xu W, Lei JJ. 2018. Creating interspecific hybrids with improved cold resistance in Fragaria. Sci Hortic. 234:1–9.
- Stamatakis A. 2014. RAxML Version 8: a tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 30:1312–1313.
- Sun J, Sun R, Liu HB, Chang LL, Li ST, Zhao MZ, Shennan C, Lei JJ, Dong J, Zhong CF, Xue L, et al. 2021. Complete chloroplast genome sequencing of ten wild Fragaria species in China provides evidence for phylogenetic evolution of Fragaria. Genomics. 113:1170–1179.
- The CAS Editorial Committee of the Flora of China. 1985. The flora of China. Vol. 37. Beijing: Science Press.