Abstract
Ormosia is a particular genus in the Fabaceae family with its striking seeds. The genus Ormosia boluoensis is a newly reported and critically endangered species, and field investigations have indicated that there are only hundreds of it. For the effective conservation, we report its complete mitochondrial genome. The length of the O. boluoensis mitochondrial genome is 248,619 bp, including 28 protein-coding genes, 14 transfer RNA genes, 3 ribosomal RNA genes, and 45 simple sequence repeats. Phylogenetic analysis revealed that O. boluoensis was a sister to the clade including Sophora flavescens, Ammopiptanthus nanus, and Ammopiptanthus mongolicus.
Fabaceae is the third largest family among the angiosperms, including more than 700 genera and 20,000 species. The Ormosia is a monophyletic but small genus in the Fabaceae family. It comprises approximately 130 species (Liu et al. Citation2019) and China contains 39 of them (http://www.iplant.cn/info/Ormosia). The Ormosia species are striking for their seeds, black or red, used as decorations, such as jewelry, beads and other trinkets. Ormosia boluoensis Y. Q. Wang & P. Y. Chen is a newly reported species (Wang and Chen Citation1995). Phylogenetic analysis based on chloroplast genomes indicated that it is genetically close to O. formosana (Wang et al. Citation2020). Unlike some widely distributed congeneric species, O. boluoensis is only found in Guangdong Xiangtoushan National Natural Reserve, China, with hundreds of individuals (Guo et al. Citation2020). It is considered an endangered species. Mitochondria are important intracellular organelles for cellular respiration and metabolism (Mackenzie and McIntosh Citation1999) and are also useful for taxonomic studies (Duminil and Besnard Citation2021). Therefore, we sequenced and reported the mitochondrial genome of O. boluoensis as a genomic resource for better conservation of this precious species.
Fresh leaves of O. boluoensis were obtained from the Guangdong Xiangtoushan National Natural Reserve, Huizhou City, China (23°16′44″N, 114°22′26″E). A voucher specimen was deposited at the Herbarium of South China Botanical Garden (Fei-Yan Zeng, [email protected]) with No. IBSC0000922. The genomic DNA of O. boluoensis was extracted using a modified CTAB method. The extracted DNA was stored in the Ecological Genetics Laboratory in South China Botanical Garden. The isolated DNA were constructed as a long- and a short-read library. The two libraries were then sequenced using the Nanopore promethION and Illumia HiSeq X Ten platforms. Before O. boluoensis mitochondrial genome assembly, minimap2 v2.17-r974-dirty (Li Citation2018) was used to align both the short and long reads to the mitochondrial genomes in Fabaceae () and the mapped reads were then extracted. Flye 2.8.1-b1676 (Kolmogorov et al. Citation2019) was subsequently used to de novo assemble the O. boluoensis mitochondrial genome with the extracted long reads. After assembly, the mitochondrial genome was polished by Racon v1.4.13 (Vaser et al. Citation2017) using the long reads and nextPolish v1.2.3 (Hu et al. Citation2020) using the short reads, respectively, each with two runs. The polished genome was annotated with GeSeq (Tillich et al. Citation2017). The simple sequence repeat (SSR) was identified with MISA-web (Beier et al. Citation2017). After annotation, the assembled O. boluoensis mitochondrial genome and its annotated files were submitted to GenBank (the accession number MW455117). To perform phylogenetic analysis for O. boluoensis, the mitochondrial genomes of the other 18 species including 16 Fabaceae species and two outgroup species (Nicotiana attenuate and Nicotiana tabacum) were downloaded from GenBank. 20 shared protein coding genes in their mitochondrion genomes were then extracted, translated into amino acid sequence, aligned and concatenated using PhyloSuite 1.2.2 (Zhang et al. Citation2020). IQ-TREE 2.1.2 (Nguyen et al. Citation2015) was used to perform maximum likelihood phylogenetic inference and the JTTDCMut model was selected as best-fit model by ModelFinder (Kalyaanamoorthy et al. Citation2017).
Figure 1. Phylogenetic tree for Ormosia boluoensis and the other species using their complete mitochondrial genomes. The GenBank accession numbers of species are shown in parentheses. Bootstrap support values in % are shown at nodes.
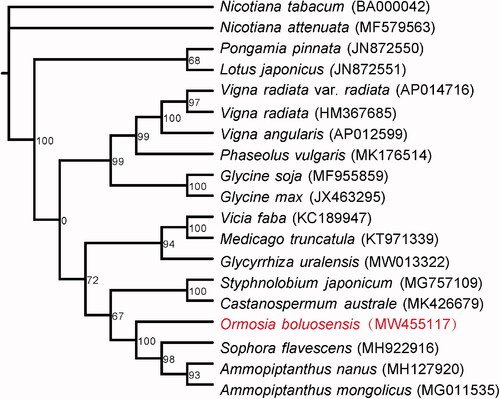
The mitochondrial genome of O. boluoensis was 248,619 bp in length. Its GC content was 45.22%. The mitochondrial genome included 28 protein-coding genes, 14 transfer RNA genes, and 3 ribosomal RNA genes. SSR analysis indicated that O. boluoensis mitochondrial genome contained 45 SSRs (Table S1), in which mononucleotide-repeat SSRs were the most abundant with 25 SSRs, followed by pentanucleotide-repeat SSRs with 11 SSRs. Phylogenetic analysis supported a monophyletic relationship of O. boluoensis to the clade of Sophora flavescens, Ammopiptanthus nanus, and Ammopiptanthus mongolicus ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete mitochondrial genome sequences of Ormosia boluoensis have been deposited in GenBank under the accession number MW455117 and is also accessible at https://doi.org/10.13140/RG.2.2.26491.95523. The associated BioProject and Bio-Sample numbers are PRJNA689914, SAMN17221877, SRA for short reads and long reads are SRR13364204 and SRR13364203 respectively.
Additional information
Funding
References
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33:2583–2585.
- Duminil J, Besnard G. 2021. Utility of the mitochondrial genome in plant taxonomic studies. In: Besse P, editor. Molecular plant taxonomy. Methods in molecular biology, vol. 2222, pp. 107-118. New York: Humana.
- Guo Y, Wang Z-F, Cao H-L. 2020. The complete chloroplast genome sequence of Ormosia boluoensis. Mitochondrial DNA Part B. 5(1):999–1000.
- Hu J, Fan J, Sun Z, Liu S. 2020. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 36:2253–2255.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546.
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100.
- Liu H, Su Z, Yu S, Liu J, Yin X, Zhang G, Liu W, Li B. 2019. Genome comparison reveals mutation hotspots in the chloroplast genome and phylogenetic relationships of Ormosia species. BioMed Res Int. 2019:7265030.
- Mackenzie S, McIntosh L. 1999. Higher plant mitochondria. Plant Cell. 11:571–585.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-T REE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution, 32:268–274. https://doi.org/10.1093/molbev/msu300.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27:737–746.
- Wang Y, Chen P. 1995. New taxa of Guangdong plants. J Tropical Subtropical Bot. 3(1):29–33.
- Wang Z-F, Chang L-W, Lian J-Y, Cao H-L. 2020. The complete chloroplast genome sequence of Ormosia formosana. Mitochondrial DNA Part B. 5(3):2636–2637.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
