Abstract
The complete mitochondrial genome of the near threatened mahseer fish, Neolissochilus hexastichus, was characterized for the first-time using Ion Torrent NGS platform. The total length of the mitogenome was 16,538 bp including a standard set of 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), 13 protein-coding genes and a non-coding control region. Twenty-eight of the 37 genes are located on the light strand and, the remaining nine genes are situated on the heavy strand. Phylogenetic analysis showed the sister relationship between N. hexastichus and N. hexagonolepis. The mitogenome could be useful for phylogenetics, population genetics, and conservation of the mahseers.
Mahseers (Family: Cyprinidae; Order: Cypriniformes), one of the largest freshwater fishes of the world, are distributed in the biodiversity hot-spots of South and Southeast Asia. Currently, 54 valid mahseer fishes have been reported under four genera viz., Tor (17 numbers), Parator (1) Neolissochilus (34) and Naziritor (2) (Fricke et al. Citation2020). These large-bodied fish are excellent sport fish and also provide livelihood to the local people. However, due to several anthropogenic factors, the natural stocks of mahseer are dwindling rapidly and, the conservation status of several mahseer species has been assessed under different categories of IUCN Red list. Measures to conserve mahseers are hindered due to high phenotypic plasticity and lack of stable taxonomic features (Pinder et al. Citation2019). DNA-based taxonomy complements the traditional methods and resolves the ambiguity to provide accurate phylogenetic relationships.
Initially, Neolissochilus hexastichus was described under the genus ‘Barbus’ (McClelland 1839) as Barbus hexastichus. Later, Rainboth (Citation1985) placed this species in the genus ‘Neolissochilus.’ Afterwards, Menon (Citation1999) synonymized the species with Tor tor as they share similar morphological characters. Recently, several researchers have validated the species status and placed it in the genus ‘Neolissochilus’ using morphology and partial gene sequences (Laskar et al. Citation2013; Lalramliana et al. Citation2019). Phylogenies with large genome coverage reveal the accurate evolutionary relationship among the species. In this study, the complete mitochondrial genome of Neolissochilus hexastichus (McClelland, 1839), a ’near-threatened’ mahseer, was characterized for the first time and its phylogenetic position was investigated within the Torinae subfamily.
One specimen of N. hexastichus was collected from River Myntdu, Jowai, West Jaintia Hills, Meghalaya (geographical coordinates: 25°26′'N 92°12′'E) by angling. The voucher specimen was deposited and, preserved in Division of Fish Genetics and Biotechnology, ICAR-Central Institute of Fisheries Education, Mumbai (A. Pavan Kumar, [email protected]) under the voucher number ‘FGBSERB-NH-2028.’ The specimen was morphologically identified using the reported literature (Talwar and Jhingran Citation1991). The total genomic DNA was isolated from the fin tissue using the Phenol–Chloroform method and, the mitochondrial genome was amplified in two overlapping amplicons by long PCR using the universal primers reported by Kim et al. (Citation2004). The amplicons (∼8 kb) were fragmented to a size of 400 bp and, used for preparing DNA libraries with NEB Next Fast DNA library preparation kit following the manufacturer's guidelines. The libraries were sequenced using Ion Torrent PGMTM 314 chip (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's guidelines. High-quality reads (Q > 30) were assembled into contigs and, mapped to the mitogenome of a closely related species N. hexagonolepis (GenBank Accession no. KU380329). The mitogenome was annotated using online tool MITOS and, the sequence submitted to the NCBI GenBank with the Accession no. MN378521. To investigate the phylogenetic position of N. hexastichus within the mahseer group, reported mitochondrial genome sequences were downloaded from the NCBI GenBank and the 13 protein coding genes were concatenated for each species. The phylogenetic tree was reconstructed using the maximum-likelihood and Bayesian inference by PAUPv4 and Mr Bayes software, respectively.
The total length of the mitogenome was 16,538 bp including a standard set of 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), 13 protein-coding genes and a non-coding control region. The frequency of nucleotides is A:31.6%, T:25%, G:15.9% and C:27.5% with a GC content of 43.4%. Twenty-eight of the 37 genes are located on the light strand (G + T content: 40.89%) and, the remaining nine genes are on the heavy strand (G + T content: 59.11%). Gene overlapping was observed between tRNAs (tRNAIle–tRNAGln; tRNAThr–tRNAPro), and protein-coding genes (ND4L–ND4; ND5–ND6; ATPase8–ATPase6). Cumulatively, 35 intergenic nucleotides (range 1–15 bp) were dispersed at 12 locations on the genome. Among all the protein-coding genes, the longest and shortest genes were ND5 (1824 bp) and ATPase8 (165 bp), respectively. Twelve of the 13 protein-coding genes used the start codon ‘ATG,’ and the COI gene used ‘GTG’ as the initiation codon. Five protein-coding genes (ND1, COI, ND4L, ND5, and ND6) showed the typical stop codon ‘TAA’. Remaining genes had incomplete stop codons ‘T- -’ (Cyt b, ND2, ND3, ND4, COII) or ’TA–’ (ATPase6, COIII). A total of 3803 codons were identified, including all the protein-coding genes. Highest numbers of codons were observed for leucine amino acid. The size of the tRNA genes varied from 66 (tRNACys) to 76 bp (tRNALeu & tRNALys). Apart from 20 tRNA genes coding for 20 amino acids, two additional tRNA genes are observed for serine (gcu) and leucine (uag) amino acids. The length of 12S and 16S rRNA genes is 956 and 1679 bp, respectively.
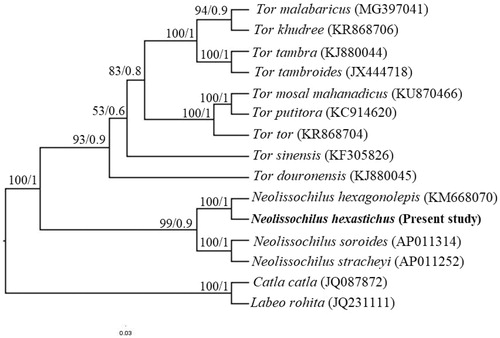
In the consensus phylogenetic tree, species of Tor and Neolissochilus formed into two distinct clades (). Within Neolissochilus clade, N. hexastichus appeared as sister species of N. hexagonolepis with a significant bootstrap value. The mitochondrial genome of N. hexastichus would be useful for conservation, population, and phylogenetics.
Acknowledgment
The first author acknowledges the ICAR, New Delhi, for providing the institutional fellowship to carry out the present study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov) under the accession number of MN378521.
Additional information
Funding
References
- Fricke R, Eschmeyer WN, van der Laan R. 2020. Eschmeyer’s catalog of fishes: genera, species, references. (http://researcharchive.calacademy.org/research/ichthyology/-catalog/.fishcatmain.asp). Electronic version accessed on 04-11-2020.
- Pinder AC, Britton JR, Harrison AJ, Nautiyal P, Bower SD, Cooke SJ, Lockett S, Everard M, Katwate U, Ranjeet K, et al. 2019. Mahseer (Tor spp.) fishes of the world: status, challenges and opportunities for conservation. Rev Fish Biol Fisheries. 29(2):417–452.
- Menon AGK. 1999. Check list – fresh water fishes of India. Records of the Zoological Survey of India. Occasional Paper No. 175:i–xxix– +1–366.
- Laskar BA, Bhattacharjee MJ, Dhar B, Mahadani P, Kundu S, Ghosh SK. 2013. The species dilemma of Northeast Indian Mahseer (Actinopterygii: Cyprinidae): DNA barcoding in clarifying the riddle. PLoS One. 8(1):e53704–e53711.
- Lalramliana , Lalronunga S, Kumar S, Singh M. 2019. DNA barcoding revealed a new species of Neolissochilus Rainboth, 1985 from the Kaladan River of Mizoram, North East India. Mitochondrial DNA Part A. 30(1):52–59.
- Kim I-C, Kweon H-S, Kim YJ, Kim C-B, Gye MC, Lee W-O, Lee Y-S, Lee J-S. 2004. The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene. 336(2):147–153.
- Rainboth WJ. 1985. Neolissochilus, a new genus of South Asian cyprinid fishes. Beaufortia. 35 (3):25–35.
- Talwar PK, Jhingran AG. 1991. Inland fishes of India and adjacent countries, 2 vols. New Delhi, Bombay, Calcutta: Oxford & IBH Publishing Co. v. 1–2: i–xvii + 36 unnumbered + 1–1158, 1 pl, 1 map. [V. 1, i–liv + 1–541, 1 map; v. 2, i–xxii + 543–1158, 1 pl].