Abstract
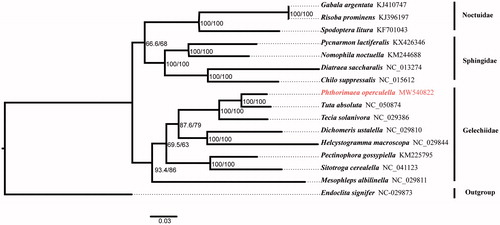
In this research, the complete mitochondrial genome (mitogenome) of Phthorimaea operculella was sequenced and annotated. The mitogenome of P. operculella is 15,269 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosome RNA (12s and 16srRNA) genes and 1 control region. In addition, we used Endoclita signifier as the outgroup to analyze phylogenetic relationship, and the phylogenetic tree showed the sister relationship between P. operculella and Tuta absoluta.
Potato tuberworm, Phthorimaea operculella (Lepidoptera: Gelechioidea: Gelechiidae), is a major pest of Solanaceae crops (Rondon Citation2010). This pest spreads all over the world especially subtropic, tropical, and Mediterranean regions, including China. It is difficult to control this pest because the female adults usually lay eggs in hiding places, such as plant tuber and foliage, meanwhile, the larvae can survive in both field and storage condition (Sporleder et al. Citation2004; Yuan et al. Citation2018). The larvae of P. operculella mainly feed on Solanaceae crops like potato, tobacco, and tomato. In China, the loss of average potato yield can reach 40–45% (Rondon and Gao Citation2018), which makes a considerable damage to the industry of potato. Previous studies mainly focused on the ecology, biology, and management of P. operculella, yet few people studied its molecular characteristic. Consequently, we determined to sequence the mitogenome of P. operculella to provide the fundamental information about its genome.
The larvae were collected from Puding county (105˚27′49″E, 26˚26′36″N), Guizhou, China, in November 2020 and reared in the laboratory of the Institute of Entomology, Guizhou University, Guiyang, China. The voucher specimen’s genome DNA and genitalia are deposited in the Institute of Entomology, Guizhou University, and the sample number is GUGC-Pht-00203 (Jian-Feng Liu, [email protected]). We used Qiagen DNeasy Blood and Tissue Kit (Cat. no. 69504) to extract the total DNA from a second instar larvae of P. operculella. The Illumina ReSeq library was prepared with an average insert size of 400 bp with 150 bp paired-end and then we used the Illumina Novaseq6000 platform (Berry Genomics, Beijing, China) to sequence it. To assemble and annotate complete mitogenome sequence, we used NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2017) with K-mer value and MitoZ v2.3 (Meng et al. Citation2019) with default set, respectively. Finally, the annotation was adjusted by MITOS2 (Bernt et al. Citation2013) and Geneious Prime v2020.2.4 (Kearse et al. Citation2012).
The mitogenome of P. operculella (accession no. MW540822) is 15,269 bp in length, containing 13 PCGs, 22 tRNA genes, 2 rRNA genes, and 1 control region. The content of A + T is distinctly higher than that of G + C (A:39.5%; G:7.9%; C:11.5%; T:41.0%), which is similar to other Gelechiidae mitogenome (Yuan et al. Citation2019). The 16sRNA gene and 12sRNA gene is 1341bp and 808 bp long, respectively. Most of protein-coding genes initiated with ATN, and ended with TAA, except for COX1 used CGA as the start codon and COX1, COX2, ND5 used incomplete T as the stop codon. The length of 22 tRNA ranged from 64 bp (tRNA-T) to 71 bp (tRNA-K).
To figure out the evolutionary status of P. operculella, we downloaded 15 complete mitogenomes from GenBank database. Among which, Endoclita signifier was selected as the outgroup. The nucleotide sequence of 13 PCGs (remove the third codon position) and 2 rRNA genes were aligned using MAFFT v7.394 (Katoh and Standley Citation2013) with L-INS-I algorithm, and then poorly aligned results were removed by trimAl v1.4.1 (Capella-Gutiérrez et al. Citation2009). Under the best fitting GTR + I + G model, the phylogenetic tree was created from the maximum-likelihood method using the IQTREE v1.6.3 software (Nguyen et al. Citation2015). The result showed that P. operculella clustered with other seven Gelechiidae species, and it was part of the Gelechiidae ().
Acknowledgment
The authors thank Chang Han, Likun Zhong, Hongli He, and Qian-Fen Zheng (Guizhou University) for technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Date availability statement
The data that support the findings of this study are openly available in NCBI at https:www.ncbi.nlm.nih.gov/, reference numbers KJ410747, KJ396197, KF701043, KX426346, KM244688, NC_013274, NC_015612, MW540822, NC_050874, NC_029386, NC_029810, NC-029844, KM225795, NC_041123, NC_029811, and NC_029873. The associated BioProject, BioSample, and SRA numbers are PRJNA699969, SAMN17817425, and SRR13741677, respectively (https://www.ncbi.nlm.nih.gov/sra/?term=SRR13741677).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet E. 69(2):313–319.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markovitz S, Duran C, et al. 2012. Geneius basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. Mitoz: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol E. 32(1):268–274.
- Rondon SI. 2010. The potato tuberworm: a literature review of its biology, ecology, and control. Am J Pot Res. 87(2):149–166.
- Rondon SI, Gao Y-L. 2018. The journey of the potato tuberworm around the world. In Perveen K, editor. Moths – pests of potato, maize and sugar beet. London (UK): IntechOpen.
- Sporleder M, Kroschel J, Gutierrez Quispe MR, Lagnaoui A. 2004. A temperature-based simulation model for the potato tuberworm, Phthorimaea operculella Zeller (Lepidoptera; Gelechiidae). Environ Entomol. 33(3):477–486.
- Yuan H-G, Wu S-Y, Lei Z-R, Rondon SI, Gao Y-L. 2018. Sub-lethal effects of Beauveria bassiana (Balsamo) on field populations of the potato tuberworm Phthorimaea operculella Zeller in China. J Integr Agr. 17(4):911–918.
- Yuan M, Yang H, Dai R-H. 2019. Complete mitochondrial genome of Sitotroga cerealella (Insecta: Lepidoptera: Gelechiidae). Mitochondrial DNA Part B. 4(1):235–236.