Abstract
Copper shark (Carcharhinus brachyurus Günther, 1870) is one of the most widely distributed but least known species in the family Carcharhinidae. Herein, we report the first complete mitogenome of C. brachyurus. The overall structure of the 16,704 bp C. brachyurus mitogenome was similar to that of other Carcharhinus species and showed the highest average nucleotide identity (97.1%) with the spinner shark (Carcharhinus brevipinna). Multigene phylogeny using 13 protein-coding genes (PCGs) in the mitogenome resolved C. brachyurus clustered with other species within the genus; the overall tree topology was congruent with recent phylogenetic studies of this species. These results provide important information for conservation genetics and further evolutionary studies of sharks.
Copper shark (Carcharhinus brachyurus Günther, 1870), in the family Carcharhinidae, is distributed in the equatorial temperate region of the eastern Atlantic, northwestern and eastern Pacific, southern Africa, Australia, and New Zealand (Compagno et al. Citation2005; Kim et al. Citation2019). In the IUNC Red List of threatened species, C. brachyurus is classified as ‘vulnerable’ (Huveneers et al. Citation2020). Various traits, including slow growth and late maturity render them vulnerable to anthropogenic activities (Drew et al. Citation2017). In the mitogenome of C. brachyurus, only the mitochondrial control region has been partially sequenced and used for phylogeographic analysis (Benavides et al. Citation2011). In this study, we analyzed the complete mitogenome sequence of C. brachyurus.
The specimens of C. brachyurus used in this study were collected from the Moseulpo fish markets in Jeju, Korea, in October 2017, as previously described (Kim et al. Citation2019). Muscle tissue samples were collected from fresh carcasses of adult female fish caught from the sea area between Gapa Island and Mara Island (33.147N, 126.269E). The collected specimens were transported and deposited at the College of Veterinary Medicine and Research Institute for Veterinary Science, Seoul National University (https://vet.snu.ac.kr/en, Prof. Se Chang Park, [email protected]) under voucher number SNU-MO-0005.
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen Korea Ltd., Seoul, Korea) according to the manufacturer’s instructions and subjected to direct PCR-based sequencing. Fifteen primer pairs, designed from the mitogenome of a related species in the family Carcharhinidae, the spinner shark (Carcharhinu brevipinna, NC_027081.1), were used to amplify the C. brachyurus mitogenome. All PCR primers and amplification conditions are listed in Supplementary Table 1. The obtained partial sequences of C. brachyurus were assembled using Geneious R11.1.5 (Kearse et al. Citation2012), and the final complete mitogenome was annotated as previously described (Kim et al. Citation2017).
The complete C. brachyurus mitogenome (MT995631) was 16,704 bp long, with a 61.7% A + T content. It consisted of a typical set of 37 genes (2 rRNAs, 22 tRNAs, and 13 protein-coding genes [PCGs]), and the overall structure was similar to the mitogenomes of other sharks in Carcharhinidae available in MitoFish (http://mitofish.aori.u-tokyo.ac.jp/). A putative D-loop (1061 bp, 66.4% A + T content) was located between tRNAPro and tRNAPhe, and the replication origin (35 bp) was located on the H-strand between tRNAAsn and tRNACys (Supplementary Table 2). The two rRNAs were 955 bp (12S rRNA) and 1676 bp (16S rRNA) long and separated by tRNAVal. The size of the 22 tRNAs varied from 67 bp (tRNASer and tRNACys) to 75 bp (tRNALeu), with a total length of 1550 bp. Except for ND6, all PCGs were encoded on the H-strand of the genome. The 12 PCGs had the typical ATG initiation codon, whereas GTG was the initiation codon in COX1. Four types of stop codons were detected: TAA (ND1, COX1, ATP8, COX3, ND4L, and ND5), AGG (ND6), and the incomplete stop codons TA- (ATP6 and CYTB) and T– (ND2, COX2, ND3, and ND4) (Supplementary Table 2).
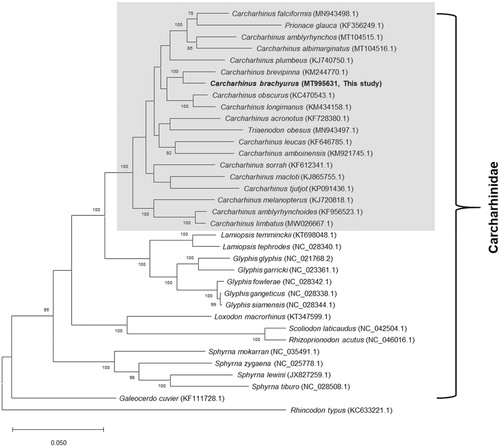
Orthologous average nucleotide identity (OrthoANI) values were analyzed with other related sharks in the family Carcharhinidae as previously described (Kim et al. Citation2017). The overall OrthoANI values between C. brachyurus and other sharks in the genus Carcharhinus were >93%, with the highest value (97.1%) obtained for spinner shark (C. brevipinna, KM244770); other genera in the family showed <92% identity (Supplementary Table 3). Thirty-three complete mitogenomes of other related species in Carcharhinidae were obtained from GenBank and used for multigene phylogenetic analysis. The concatenated sequences of the 13 PCGs were aligned as previously described (Kim et al. Citation2017), and the maximum likelihood tree was reconstructed using MEGAX version 10.0 (Kumar et al. Citation2018). In the resultant tree, copper shark was clustered with other Carcharhinus species (). Our tree topology was congruent with previous phylogenetic analyses based on nuclear or mitochondrial genes, which clustered blue shark (Prionace glauca) and whitetip reef shark (Triaenodon obesus) with other sharks in the genus Carcharhinus (Dosay-Akbulut Citation2008; Li et al. Citation2016), indicating that taxonomic assignment of the two species warrants further reevaluation. The mitogenome and associated genomic data of C. brachyurus provide important insights into biodiversity and address phylogenetic relationships within the genus Carcharhinus.
Figure 1. Maximum-likelihood phylogenetic tree based on the concatenated sequences of 13 protein-coding genes from the available Carcharhinidae mitogenomes. The gray box denotes the species currently placed in the genus Carcharhinus and other related taxa, including Prionace glauca and Triaenodon obesus. Mitochondrial genome of whale shark (Rhincodon typus, KC633221.1) was used as outgroup. Numbers at the branches indicate bootstrapping values obtained with 1,000 replicates, and only bootstrap values >70% are indicated. The scale bar represents 0.05 nucleotide substitutions per site.
Supplemental Material
Download Zip (47.2 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in the NCBI GenBank database at https://www.ncbi.nlm.nih.gov, reference number MT995631.
Additional information
Funding
References
- Benavides MT, Feldheim KA, Duffy CA, Wintner S, Braccini JM, Boomer J, Huveneers C, Rogers P, Mangel JC, Alfaro-Shigueto J, et al. 2011. Phylogeography of the copper shark (Carcharhinus brachyurus) in the southern hemisphere: implications for the conservation of a coastal apex predator. Mar Freshwater Res. 62(7):861–869.
- Compagno LJV, Dando M, Fowler S. 2005. Sharks of the world. Princeton (NJ): Princeton University Press; p. 292–293. ISBN 978-0-691-12072-0.
- Dosay-Akbulut M. 2008. The phylogenetic relationship within the genus Carcharhinus. C R Biol. 331:500–509.
- Drew M, Rogers P, Huveneers C. 2017. Slow life-history traits of a neritic predator, the bronze whaler (Carcharhinus brachyurus). Mar Freshwater Res. 68(3):461–472.
- Huveneers C, Rigby CL, Dicken M, Pacoureau N, Derrick D. 2020. Carcharhinus brachyurus. The IUCN red list of threatened species 2020: e.T41741A2954522. https://dx.doi.org/10.2305/IUCN.UK.2020-3.RLTS.T41741A2954522.en. (Accessed on March 2021).
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649.
- Kim JH, Lee YR, Koh JR, Jun JW, Giri SS, Kim HJ, Chi C, Yun S, Kim SG, Kim SW, et al. 2017. Complete mitochondrial genome of the beluga whale Delphinapterus leucas (Cetacea: Monodontidae). Conservation Genet Resour. 9(3):435–438.
- Kim SW, Han SJ, Kim Y, Jun JW, Giri SS, Chi C, Yun S, Kim HJ, Kim SG, Kang JW, et al. 2019. Heavy metal accumulation in and food safety of shark meat from Jeju Island, Republic of Korea. PLoS One. 14(3):e0212410.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li W, He S, Tian S, Dai X. 2016. Phylogeny analysis of complete mitochondrial DNA sequences for pelagic fishes from tuna fishery. Mitochondrial DNA Part B. 1(1):811–814.
