Abstract
The Red Keelback (Pseudagkistrodon rudis Boulenger, 1906) is widely distributed in the southern of China. The complete mitochondrial genome (mitogenome) of P. rudis was determined for the first time by using next-generation sequencing. The size of assembled mitogenome for P. rudis was 19,150 bp, which included 13 protein coding genes (PCGs), 22 tRNAs, two rRNAs and two control regions (d-loop1 and d-loop2). The Bayesian tree showed that P. rudis and Rhabdophis tigrinus have a closed relationship. These results can provide data for phylogeny and molecular classification of the genus.
The Red Keelback (Pseudagkistrodon rudis Boulenger, 1906) belongs to the snake family of Colubridae, which is an aggressive but harmless snake, and widely distribution in the southern of China (Uetz et al. Citation2020). The genus Macropisthodon includes two branches, the branch represented by P. rudis is an independent monotypic genus and P. rudis is a Chinese endemic species, therefore this species in China use Pseudoagkistrodon as genus name instead of Macropisthodon (Takeuchi et al. Citation2018). And P. rudis was included in the ‘National Protected List of Terrestrial Wild Animals with Good Benefits or Important Economic and Scientific Values’ issued by the State Forestry Administration of China on 1 August 2000. Mitochondrial genome (mitogenome) is widely used in phylogenetic and evolutionary studies in these days (Crimi and Rigolio Citation2008). It is the first time to study the complete mitochondrial genome of P. rudis.
The specimen of P. rudis (species voucher: LSU20200722WF01) was collected at Xiayangxiang (N27.87354294°, E119.82021360°) from Wencheng County, Wenzhou City, Zhejiang Province, China. The collected specimen was stored in 90% ethanol and deposited at the Museum of Laboratory of Amphibian Diversity Investigation (contact person: Guo-Hua Ding, E-mail: [email protected]) at Lishui University. Whole genomic DNA was extracted from muscle tissue using the EasyPure genomic DNA kit (TransGen Biotech Co., Beijing, China). The whole genome sequencing (WGS) was implemented by Illumina NovaSeq 6000 platform (Novogene Bioinformatics Technology Co. Ltd., Tianjin, China) for paired-end 150 bp. We used NOVO Plasty 3.7 (Dierckxsens et al. Citation2017) to assemble the mitogenome referencing random partail mitogenome (about 200 bp) of a snake from family Colubridae based on the WGS data, and then used MITOS WebServer (Bernt et al. Citation2013) to annotate and locate the genes.
The complete mitogenome of P. rudis (GenBank accession no. MW327508) is 19,150 bp in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), two ribosomal RNA genes (rRNA) and two control regions (d-loop1 and d-loop2). In these 37 mitochondrial genes, most of them were in the positive strand, except ND6, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro, which were in the minus chain. Among the 13 PCGs, the longest gene is ND5 (1782 bp), while the shortest is ATP8 (165 bp). The 12S rRNA (922 bp) was located between tRNAPhe and tRNAVal, and the16S rRNA (1448 bp) was located between tRNAVal and ND1. D-loop1 (1926 bp) was located between tRNAIle and tRNALeu; d-loop2 (2164 bp) was located between tRNAPro and tRNAPhe. ATA and ATG are the start codons of PCGs; only three PCGs (ND1, ND3, COI) use ATA; the others use ATG. There are six types of termination codons, include T (ND1, COIII and Cytb); TA (ATP6); TAG (ND2 and ND6); AGA (COI); TAA (COII, ATP8, ND3, and ND4L); AGG (ND4 and ND5).
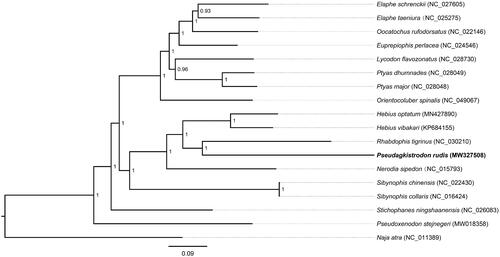
In the study, we used PhyloSuite v1.2.1 (Zhang et al. Citation2020) to assemble the 13 PCGs (11,319 bp) in mitogenomes of 18 snakes from the family of Colubridae and Elapidae. Based on the assembled PCGs the phylogenetic relationship were constructed by MrBayes v3.2.2. (Ronquist and Huelsenbeck Citation2003) (). Naja atra (Squamata: Elapidae), which is relatively close to their ancestors, was selected as the outgroup. GTR + I + G as the best-fit substitution model was obtained using MrModelTest 2.3 (Nylander Citation2004). It appeared that P. rudis and Rhabdophis tigrinus have a closed relationship, and both species were closed with Hebius species. It is similar to a previous study (Pyron et al. Citation2013). These results can provide data for phylogeny and molecular classification of the genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome data supporting this study are openly available in GenBank at (https://www.ncbi.nlm.nih.gov/nuccore/MW327508). Reference number (Accession no. MW327508). BioSample and SRA accession numbers are https://www.ncbi.nlm.nih.gov/biosample/SAMN17167287 and https://www.ncbi.nlm.nih.gov/sra/SRR13309610, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Crimi M, Rigolio R. 2008. The mitochondrial genome, a growing interest inside an organelle. Int J Nanomed. 3:51–57.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 93:1471–2148.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Takeuchi H, Savitzky AH, Ding L, de Silva A, Das I, Nguyen TT, Tsai TS, Jono T, Zhu GX, Mahaulpatha D, Tang Y, et al. 2018. Evolution of nuchal glands, unusual defensive organs of Asian natricine snakes (Serpentes: Colubridae), inferred from a molecular phylogeny. Ecol Evol. 8(20):10219–10232.
- Uetz P, Freed P, Hošek J. 2020. The reptile database [accessed 2020 December 30]. http://www.reptile-database.org.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.