Abstract
Sarcophaga tsinanensis (Fan, 1964) (Diptera: Sarcophagidae), a species from subgenus Heteronychia. In this study, we report the complete mitochondrial genome (mitogenome) of S. tsinanensis. The length of this mitogenome was 14,972 bp in total, containing 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs, and a non-coding control region. The arrangement of genes was the same as that of ancestral metazoan. Sarcophaga tsinanensis has a high nucleotide bias in A/T accounting for 76.80% of total nucleotides (A 39.9%, G 9.2%, C 13.9%, and T 36.9%). The result of phylogenetic analysis revealed that S. tsinanensis cluster together with species from the same subgenus Heteronychia, showing a clear monophyletic relationship. Sarcophaga tsinanensis is closely related to its sister species Sarcophaga depressifrons, Sarcophaga plotnikovi, and Sarcophaga shnitnikovi. This study provides the mitochondrial data of S. tsinanensis for further research on evolutionary relationships and species identification of flesh flies.
Sarcophagidae is a family of flies with forensic significance, which can provide valuable data for the estimation of minimum postmortem interval (PMImin) and other crime scene related information (Ren et al. Citation2018). Subgenus Heteronychia was the largest lineage of Sarcophaga (Diptera: Sarcophagidae) in species number (Daniel et al. Citation2013). At present, only a few species from subgenus Heteronychia has been discovered and studied (Krčmar et al. Citation2019; Ren et al. Citation2020). In this study, we presented the complete mitochondrial genome (mitogenome) of Sarcophaga tsinanensis (Fan 1964), which could further enrich our understanding of the phylogenetic relationship and phylogeography of subgenus Heteronychia.
In our study, adult specimens of S. tsinanensis were first trapped by decomposing pig livers in July 2020 in Beijing city (40°22′N, 116°23′E), China. All specimens were processed by freezing, and then identified based on traditional morphological features (Xu and Zhao 1996). All specimens were stored at −80 °C in Dr. Wang’s Lab (Changsha, Hunan, China) with a unique code (CSU20200713). Total DNA was extracted from thoracic muscle tissues using TIANamp Micro DNA Kit (Tiangen Biotech Co., Ltd, Beijing, China) according to the manufacture’s instruction. The sequencing of S. tsinanensis mitogenome was carried out with an Illumina HiSeq 2500 Platform (paired-end 150 bp) at OE Biotech. Co., Ltd. (Shanghai, China). de novo assembly was performed using MITObim v1.9 and SOAPdenovo v2.04 (https://github.com/chrishah/MITObim and http://soap.genomics.org.cn/soapdenovo.html) (Hahn et al. Citation2013). Finally, the preliminary annotation of all genes was determined by MITOS2 Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). To ensure the accuracy of the annotation, 13 protein coding genes (PCGs) were compared with published Sarcophagidae mitoticgenomes using MEGA X (Kumar et al. Citation2018), The transfer RNAs (tRNAs) were predicted by tRNAscan-SE Search Server v1.21 (Lowe and Chan Citation2016) and verified by comparison with those from other dipteran insects.
The length of S. tsinanensis mitogenome was 14,972 bp in total (GenBank no. MW423621), containing 13 PCGs, 2 ribosomal RNAs (rRNAs), 22 tRNAs, and a non-coding control region. The arrangement of genes was the same as those of ancestral metazoan (Cameron Citation2014). Among them, 9 PCGs and 14 tRNAs were encoded by H-strand, and the remaining 4 PCGs, 8 tRNAs and 2 rRNAs were encoded by L-strand. There was highly A/T bias in nucleotide composition, accounting for 76.80% of total nucleotides (A 39.9%, G 9.2%, C 13.9%, and T 36.9%). Maximum-likelihood (ML) analysis was performed in IQ-TREE v.1.6.8 with ultrafast likelihood bootstrap set to 1000 replicates (Nguyen et al. Citation2015). Phylogenetic analysis of S. tsinanensis and other 20 flesh flies were conducted using the ML method based on 13 PCGs of mitogenome, with Chrysomya pinguis (Diptera: Calliphoridae) as an outgroup (). These 20 flesh fly species come from three genera. Two species belong to Oxysarcodexia genera, two species belong to Boettcheria genera, and all other species come from the same subgenus where S. tsinanensis belongs. All these sequences were aligned using MAFFT version 7 software (Katoh and Standley Citation2013).
Figure 1. Phylogenetic trees of S. tsinanensis with other 20 flesh flies based on 13 protein-coding genes using the maximum-likelihood method. Chrysomya pinguis was selected as an outgroup.
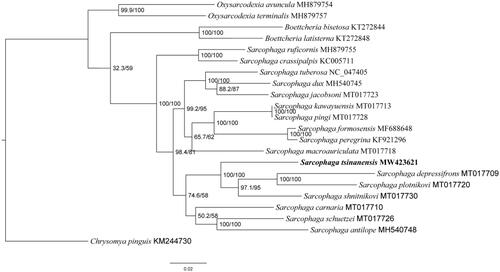
The result of phylogenetic analysis revealed that S. tsinanensis clustered together with species from the same subgenus Heteronychia, showing a clear monophyletic relationship, and S. tsinanensis is closely related to its sister species Sarcophaga depressifrons, Sarcophaga plotnikovi, and Sarcophaga shnitnikovi. Since there are only limited mitogenomes of Heteronychia subgenus currently available, this study provides mitochondrial data of S. tsinanensis for further research on evolutionary relationships and species identification of flesh flies.
Acknowledgments
The authors are grateful to Prof. Lushi Chen (Guizhou Police Officer College) for species identification.
Disclosure statement
The authors have declared no competing interest.
Data availability statement
The assembled mitochondrial genome is available on NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW423621 (Accession no. MW423621). Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA690669/, https://www.ncbi.nlm.nih.gov/sra/SRR13441657, and https://www.ncbi.nlm.nih.gov/biosample/SAMN17319966/, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117.
- Daniel W, Thomas P, Pierfilippo C. 2013. Phylogeny of Heteronychia: the largest lineage of Sarcophaga (Diptera: Sarcophagidae). Zool J Linn Soc. 169(3):604–639.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Krčmar S, Whitmore D, Pape T, Buenaventura E. 2019. Checklist of the Sarcophagidae (Diptera) of Croatia, with new records from Croatia and other Mediterranean countries. ZK. 831:95–155.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35(6):1547–1549.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ren L, Shang Y, Chen W, Meng F, Cai J, Zhu G, Chen L, Wang Y, Deng J, Guo Y. 2018. A brief review of forensically important flesh flies (Diptera: Sarcophagidae). Forensic Sci Res. 3(1):16–26.
- Ren L, Zhang X, Li Y, Shang Y, Chen S, Wang S, Qu Y, Cai J, Guo Y. 2020. Comparative analysis of mitochondrial genomes among the subfamily Sarcophaginae (Diptera: Sarcophagidae) and phylogenetic implications. Int J Biol Macromol. 161:214–222.
- Xu WQ, Zhao JM. 1996. Flies of China. Shenyang: Liaoning Science and Technology.
