Abstract
Rhus punjabensis var. sinica belongs to the family Anacardiaceae in the order Sapindales. In this study, we first reported the complete chloroplast genome sequence of R. punjabensis var. sinica. The cp genome was sequenced on Illumina Hiseq 2000 platform. The sequence was found to be 159,617 bp in length with 37.9% GC contents, including a large single-copy region of 87,694 bp, a small single-copy region of 18,971 bp, and a pair of inverted repeats of 26,476 bp. The chloroplast genome of R. punjabensis var. sinica contains 133 genes, including 86 protein-coding genes, 8 rRNA genes, and 2 pseudogenes identified by CPGAVAS2 and BLAST search, and 37 tRNA genes annotated by tRNAscan-SE. Maximum-likelihood (ML) phylogenetic analysis showed that R. punjabensis var. sinica was sister to Rhus potaninii.
Rhus punjabensis var. sinica, belonging to the family Anacardiaceae in the order Sapindales, grows on hill and mountain forests at an altitude of 400−3000 m. Galla Chinensis, a natural traditional Chinese medicine, is formed by Rhus gall aphids that live on the leaves, petioles, and wings of the primary host plants Rhus (Ren et al. Citation2017), and is widely used in China (Zhang et al. Citation2015). The host plants include Rhus chinensis, Rhus potaninii, R. punjabensis var. sinica, Rhus typhina, and Rhus glabra. The main component of Galla Chinensis is tannic acid, which has antioxidation effect (Tajima et al. Citation2016), antidiarrheal effect (Yang et al. Citation2017), analgesic and anti-inflammatory effects (Sun et al. Citation2018). Because there are many researches related to pharmacology but less on its genome, this study provides a theoretical basis for the phylogenetic relationship of Rhus and the coevolution between host trees and Rhus gall aphids.
The specimen was stored in Herbarium of Institute of Medicinal Plant Development (voucher: Pan0102). Fresh leaves of R. punjabensis var. sinica were collected from Enshi City, Hubei Province (29°44′02″N, 109°29′48″E) at an altitude of 600 m on 5 September 2019. Its total genomic DNA was extracted using QIAGEN DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The whole genome was sequenced on Illumina Hiseq 2000 platform (Illumina, San Diego, CA), and 8.12 G data were acquired. Clean data were further assembled into a complete chloroplast genome using SOAPdenovo version 2 (Hong Kong, China) (Luo et al. Citation2012) and SSPACE (Boetzer et al. Citation2011). The protein-coding genes, rRNA genes, and pseudogenes were identified by CPGAVAS2 (Shi et al. Citation2019) and BLAST search, and tRNA genes were annotated by tRNAscan-SE (Schattner et al. Citation2005).
The chloroplast genome of R. punjabensis var. sinica (GenBank accession number: MT230555) was 159,617 bp long with 37.9% GC content. The GC content in IR regions, large-single copy (LSC) region, and small single-copy region (SSC) region were 43.0%, 36.0%, and 32.6%, respectively. The genome includes a LSC region of 87,694 bp, a SSC region of 18,971 bp, and a pair of inverted repeats of 26,476 bp. The genome contains 86 protein-coding genes, 8 rRNA genes, 2 pseudogenes, and 37 tRNA genes. In the protein-coding regions, the AT content of the third codon position (69.2%) was higher than that of the first (54.1%) and the second codon positions (61.8%).
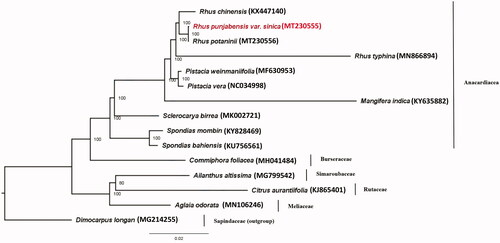
The maximum-likelihood (ML) tree (Schattner et al. Citation2005) of R. punjabensis var. sinica and 14 species from order Sapindales based on complete chloroplast genome sequence, was constructed with Dimocarpus longan as outgroup (). The ML tree with 1000 replicates revealed that the family Anacardiaceae was strongly supported as a monophyletic group and four species of Rhus were clustered into a clade. Rhus punjabensis var. sinica was sister to R. potaninii with 100% support value.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession No. MT230555. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA688107, SRR13341544, and SAMN17169251, respectively.
Additional information
Funding
References
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 27(4):578–579.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1(1):18.
- Ren Z, Harris AJ, Dikow RB, Ma E, Zhong Y, Wen J. 2017. Another look at the phylogenetic relationships and intercontinental biogeography of eastern Asian - North American Rhus gall aphids (Hemiptera: Aphididae: Eriosomatinae): evidence from mitogenome sequences via genome skimming. Mol Phylogenet Evol. 117:102–110.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Sun K, Song X, Jia R, Yin Z, Zou Y, Li L, Yin L, He C, Liang X, Yue G, et al. 2018. Evaluation of analgesic and anti-inflammatory activities of water extract of Galla chinensis in vivo models. Evid Based Complement Alternat Med. 2018:1–7.
- Tajima N, Takasaki M, Fukamachi H, Igarashi T, Nakajima Y, Arakawa H. 2016. Determination of reactive oxygen generated from natural medicines and their antibacterial activity. J Pharm Anal. 6(4):214–218.
- Yang Y, Luo H, Song X, Yu L, Xie J, Yang J, Jia R, Lin J, Zou Y, Li L, et al. 2017. Preparation of Galla chinensis oral solution as well as its stability, safety, and antidiarrheal activity evaluation. Evid Based Complement Alternat Med. 2017:1851459.
- Zhang T, Chu J, Zhou X. 2015. Anti-carious effects of Galla chinensis: a systematic rview. Phytother Res. 29(12):1837–1842.