Abstract
Vicia cracca L. is a widespread perennial herb in the Northern Hemisphere. It has purple flowers and leave tendrils for climbing on neighboring vegetation. For knowing the chloroplast genome, a sample’s genomic was extracted, sequenced, assembled and annotated. The chloroplast genome of this plant is a circular form of 126,272bp in length with IR loss. After annotation, a total of 108 genes were predicted, of which, 75 encode proteins, 3 rRNA, 30 tRNA. The evolutionary history, inferred using Maximum Likelihood method, indicates that V. cracca was grouped within Vicia in Fabaceae. The complete cp genome will be helpful for further studies on molecular biology, evolution, population genetics, taxonomy or resources protection.
Vicia cracca L., belonging to Fabaceae, is widespread in the Northern Hemisphere. It is a polycarpic perennial herb and often found in meadows, along roads, river banks, and forest margins. This plant has dense pedunculate racemes inflorescences comprising of 10–30 purple flowers and with leave tendrils, its stems can climb on neighboring vegetation (Eliášová et al. Citation2014; Eliášová and Münzbergová Citation2017). In previous studies, many literatures focused on chemical composition, genetic diversity of autotetraploids in natural populations, infecting viruses, root nodulating Rhizobium leguminosarum (Christian et al. Citation1982; Eliášová et al. Citation2014; Van Cauwenberghe et al. Citation2014; Gallet et al. Citation2018). In this study, we report the complete chloroplast (cp) genome of V. cracca., and analyzed the relationship with other related species by phylogenetic analysis.
Samples from Qilian mountains (36°34′37″N,101°48′27″E) in Qinghai province were collected for sequencing. A specimen was deposited at College of Ecological Environment and Resources, Qinghai Nationalities University (https://shxy.qhmu.edu.cn/, Junqiao Li, email: [email protected]) under the voucher number HCEERQNU-20200827001. A sample’s total genomic DNA was extracted from about 100 mg fresh leaves using a modified CTAB method (Murray and Thompson Citation1980). Paired-end Libraries with an average length of 350 bp were constructed and sequenced on the Illumina Novaseq platform (Shenzhen Huitong Biotechnology Co. Ltd). In total, 13,677,281 raw reads were obtained with 350X coverage. After filtered by NGSQCToolkit v2.3.3 (Patel and Jain Citation2012), 13,625,886 clean reads were used for assembling with the de novo assembler SPAdes (Bankevich et al. Citation2012). Gene annotation was performed via PGA (Qu et al. Citation2019).
The complete cp genome of V. cracca (GenBank accession no. MW266076) is a circular form of 126,272bp in length with 34.8% GC content and IR loss which is common in Fabaceae, especially in Papilionoideae (Cai et al. Citation2008; Yi et al. Citation2020). A total of 108 genes were predicted on this cp genome to composing of 75 encode proteins, 3 rRNA and 30 tRNA.
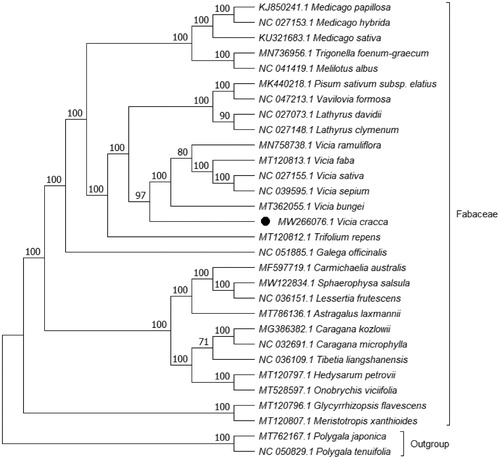
Phylogenetic analysis was performed based on complete cp genomes of V. cracca and other 27 related species reported in Fabaceae, two species in Polygalaceae as outgroup. The genome-wide alignment of 30 genomes was constructed by HomBlocks (Bi et al. Citation2018). The evolutionary history was inferred with Maximum Likelihood (ML) method by IQ-TREE 1.6.12 under TVM + F+R3 model (Nguyen et al. Citation2015; Kalyaanamoorthy et al. Citation2017), and the output file was edited in MEG X (Kumar et al. Citation2018). Bootstrap (BS) values were calculated with UFBoot2 from 1000 replicates (Hoang et al. Citation2018). As expected, V. cracca was grouped within vicia in Fabaceae (). The phylogenetic relationship was generally consistent with the result of Schaefer’s research on tribe Fabeae Systematics (Schaefer et al. Citation2012). The complete cp genome of V. cracca will be helpful for further studies on molecular biology, evolution, population genetics, taxonomy or resources protection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MW266076) under the accession no. MW266076. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA687614, SRR13300064, and SAMN17150903, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bi GQ, Mao YX, Xing QK, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Cai ZQ, Guisinger M, Kim HG, Ruck E, Blazier JC, McMurtry V, Kuehl JV, Boore J, Jansen RK. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 67(6):696–704.
- Van Cauwenberghe J, Verstraete B, Lemaire B, Lievens B, Michiels J, Honnay O. 2014. Plant population structure of root nodulating Rhizobium leguminosarum in Vicia cracca populations at local to regional geographic scales. Syst Appl Microbiol. 37(8):613–621.
- Christian M, Baumann A, Donny S, Harold R. 1982. Purification and characterization of a mannose/glucose-specific lectin from Vicia cracca. Eur J Biochem. 122(1):105–110.
- Eliášová A, Münzbergová Z. 2017. Factors influencing distribution and local coexistence of diploids and tetraploids of Vicia cracca: inferences from a common garden experiment. J Plant Res. 130(4):677–687.
- Eliášová A, Pavel T, Bohumil M, Zuzana M. 2014. Autotetraploids of Vicia cracca show a higher allelic richness in natural populations and a higher seed set after artificial selfing than diploids. Ann Bot. 113(1):159–170.
- Gallet R, Kraberger S, Filloux D, Galzi S, Fontes H, Martin DP, Varsani A, Roumagnac P. 2018. Nanovirus‑alphasatellite complex identified in Vicia cracca in the Rhône delta region of France. Arch Virol. 163(3):695–700.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35(2):518–522.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 8(19):4321–4326.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7:e30619.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Schaefer H, Hechenleitner P, Santos-Guerra A, de Sequeira MM, Pennington RT, Kenicer G, Carine MA. 2012. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evol Biol. 12(1):250.
- Yi FY, Zhan C, Wang HM, Yan XH, Ye RH, Gong Q, Qiu X, Liu QS, Sun HL. 2020. Characterization of the complete chloroplast genome sequence of Vicia costata (Fabaceae) and its phylogenetic implications. Mitochondrial DNA Part B. 5(3):3463–3464.