Abstract
Turdus ruficollis is mainly found in China and Northeast Asia. So far, the mitochondrial genome of more than 20 species from the genus Turdus has been studied. However, the relevant information of T. ruficollis has not been reported. To grasp a better comprehension on the molecular basis of T. ruficollis, we obtained the complete mitochondrial DNA genome sequences of this species. The mitogenome was 16,737 bp in length, which consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one control region. A phylogenetic tree based on complete mitogenome sequences revealed that, within the genus Turdidae, T. ruficollis is closely related to T. naumanni and T. eumomus. The complete mitochondrial genome of T. ruficollis would be of great utility for population genetics and phylogeography of the Turdidae family and would also provide meritorious insights for future studies on conservation, genetics, and phylogeny of the Passeriformes family.
Turdus ruficollis (Passeriformes: Turdidae), known as red-necked thrush, breeds in South-Central Siberia and winters in northern China (Liu et al. Citation1987; Xu et al. Citation2010; Zheng Citation2017). In this study, the sample was taken from a red-necked thrush with unknown cause of death on campus of Qinghai Nationalities University, Xining City, Qinghai Province, China (36.5874N; 101.8199E). The specimen of T. ruficollis was deposited at the animal specimen room in Herbarium of College of Ecological Environment and Resources, Qinghai Nationalities University (www.qhmu.edu.cn, Zhang Xuze, [email protected]) under the voucher number XNTR-01. Genomic DNA was extracted from the muscle tissue, using a modified method adopted from the standard phenol/chloroform extraction process (Sambrook et al. Citation1989), with digestion time extended to 12 hours and an additional round of phenol/chloroform extraction.
The DNA extract was sheared and sequenced on an Illumina NovaSeq (Illumina, San Diego, CA) following standard Illumina pair-end sequencing protocol with a read length of 150 bp. The average coverage of genome was about 5627. Raw reads were filtered with fastp (version 0.20.0, https://github.com/OpenGene/fastp) and 22,284,470 clean pair-end reads were assembled into a complete mitogenome with SPAdes (Bankevich et al. Citation2012). The whole sequence was annotated with the software Geneious v11.1.5, and tRNA genes were predicted by the online software MITOS (Bernt et al. Citation2013; Zhang et al. Citation2020).
The assembled T. ruficollis mitogenome was a closed loop of 16,737 bp, of which 11,396 nucleotides were coding DNA. The mitochondrial DNA sequence has been deposited in GenBank (accession no. MT712159). The mitogenome comprised 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and one control region. The overall base composition of the whole mitochondrial genome was 29.28% A, 23.27% T, 32.5% C, and 14.95% G, exhibiting obvious AT bias (52.55%), consistent with the typical base bias found in Turdidae mitochondria (Li et al. Citation2014; Sun et al. Citation2020).
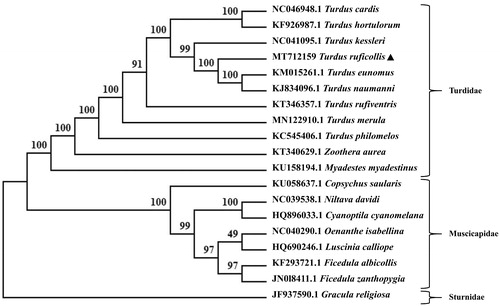
Phylogenetic relationship of T. ruficollis with 18 species of Passeriformes base on the complete sequences of mitochondrial DNA was resolved by means of maximum-likelihood (ML). The ML tree was built with MEGA X (Kumar et al. Citation2018), based on the Kimura 2-parameter model, with bootstrap set to 1000 and Gracula religiosa taken as an outgroup. The phylogenetic tree suggested that T. ruficollis and (T. naumanni + T. eumomus) were clustered into one branch with high nodal support (100) (). The species from the Turdidae family and those from the Muscicapidae family clustered into two separate branches. The present study has confirmed the consistency between morphological classification and molecular classification (Cheng Citation1987; Zheng Citation2017; Dong et al. Citation2018). The sequencing of the red-necked thrush mitochondrial genome is of particular interest for better understanding of population genetics and phylogeography of the Turdidae family. This study would also provide more basic data on conservation, genetics and phylogeny of the Passeriformes family for future research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitochondrial genome sequence in the present study is openly available at GenBank (https://www.ncbi.nlm.nih.gov/) under the accession number MT712159. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA679709, SUB8581141, and SAMN16844544, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cheng Z. 1987. A synopsis of the avifauna of China. Hamburg, Berlin/Beijing: Parey/Science Press.
- Dong Y, Li B, Zhou L. 2018. A new insight into the classification of dusky thrush complex: bearings on the phylogenetic relationships within the Turdidae. Mitochondrial DNA Part A. 29(8):1245–1248.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li B, Zhou LZ, Liu G, Gu CM. 2014. Complete mitochondrial genome of Naumann's thrush Turdus naumanni (Passeriformes: Turdidae). Mitochondrial DNA Part A. 27(2):1117–1118.
- Liu HJ, Su HL, Shen SY, Lan YT, Ren JQ. 1987. Studies on the population structure of the red-necked thrush and analysis of its food pattern during winter. J Shanxi Univ. 4:92–100.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
- Sun C-H, Xu P, Lu C-H, Han Q, Lin Y-F, Gao Z-J, Lu C-H. 2020. Complete mitochondrial genome of the Grey thrush Turdus cardis (Aves, Turdidae). Mitochondrial DNA B. 5(2):1824–1825.
- Xu HL, Xiong W, Yao YF. 2010. Phylogeny of fourteen Turdinae birds based on mitochondrial Co I gene sequences. J Northeast Forest Univ. 38(10):75–78.
- Zhang XZ, Fu L, Guo SC. 2020. The sequence and characterization of mitochondrial of Lepus oiostolus (Lagomorpha: Leporidae). Mitochondrial DNA B. 5(3):2135–2136.
- Zheng GM, editor. 2017. A checklist on the classification and distribution of the birds of China. 3rd ed. Beijing: Science Press.