Abstract
Gyrinops walla is an important agarwood-producing tree and threatened species from Sri Lanka. Herein, we assembled and characterized the complete chloroplast (cp) genome of G. walla as a genomic resource for conservation purposes. The 175,130 bp long genome is comprised of 87,376 bp large single-copy (LSC) and 3316 bp small single-copy (SSC) regions, which are separated by two inverted repeat (IR) region, each with a size of 42,291 bp. A total of 140 genes were predicted for the cp genome, which includes 94 protein-coding, 38 tRNA, and eight rRNA genes. Phylogenetic analysis showed that G. walla is fully resolved in a sister position to Aquilaria in the family Thymelaeaceae. The data provided will be useful for study on the molecular phylogenetics and evolution of Thymelaeaceae in the future.
Gyrinops walla Gaertn. 1791, locally known as ‘Walla Patta’, is naturally distributed in the low and mid elevations of the wet zone of Sri Lanka (Subasinghe et al. Citation2012). The species is mainly confined to natural forests and related vegetation, where it is associated with other native species in the lower canopy layer (Subasinghe and Hettiarachchi Citation2013). The plant is known to produce fragrant resin, called the agarwood. The agarwood is sought after as raw material for perfumery and traditional Attar (Subasinghe and Hettiarachchi Citation2013). In the wild, the tree is heavily poached and illegal agarwood hunters tend to conduct illegal felling of these trees in search of the agarwood, causing its population to decrease dramatically over time. Therefore, the Sri Lankan Government listed this species under ‘Vulnerable category’ in 2012 and banned transporting and exporting of any part of the tree or product (Subasinghe Citation2013). To date, G. walla is classified under the Appendix II of the Convention on International Trade in Endangered Species (CITES) and all international trade of this species is closely monitored (UNEP-WCMC (Comps) Citation2021). Scientific studies were widely conducted on G. walla after identifying its ability of producing agarwood resins (Subasinghe and Hettiarachchi Citation2013). However, genetic research on this species is still limited (Eurlings and Gravendeel Citation2005; Farah et al. Citation2018; Pern et al. Citation2020). In this study, we characterized the complete chloroplast (cp) genome sequence of G. walla to serve as a valuable genomic resource for the conservation effort of this important agarwood-producing plant species and to determine its evolutionary relationship to other genera classified in the Thymelaeaceae.
Total genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1990) from fresh leaves of wild G. walla collected from a forest associated vegetation in the Yagirala of Kalutara District (N06°21′48″ E80°10′08″). A voucher specimen was deposited in the Department of Forestry and Environmental Science, University of Sri Jayewardenepura, Sri Lanka (https://www.sjp.ac.lk; S.M.C.U.P. Subasinghe; [email protected]) under the collection number GWYAG01-06. Using the TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA), a 300 bp insert size genomic library was constructed and sequencing was carried out on the Illumina Novaseq platform. Approximately, 2 Gb of 150 bp paired-ends raw data were generated and NOVOPlasty 4.3 (Dierckxsens et al. Citation2017) was used for the genome assembly. The rbcL sequence of G. walla (GenBank accession number: MF443411) was designated as the seed sequence. Gene annotation was conducted using GeSeq 2.03 (Tillich et al. Citation2017) based on default parameters and manually checked for errors.
The complete cp genome sequence of G. walla (GenBank accession number: MW557323) exhibited a typical quadripartite structure and has a length of 175,130 bp. The cp genome includes a large single-copy (LSC) region of 87,376 bp, a small single-copy (SSC) region of 3316 bp, separated by a pair of 42,219 bp inverted repeat (IR) regions. A total of 140 genes were predicted, including 94 protein-coding, 38 tRNA, and eight rRNA genes. The overall GC content was 36.7%.
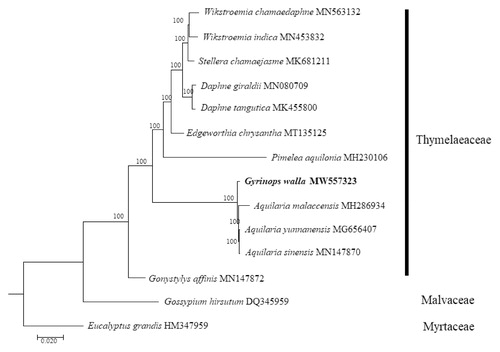
To determine the phylogenetic placement of G. walla within the family Thymelaeaceae using the complete cp genome sequences, 12 selected taxa from the family Thymelaeaceae were aligned using MAFFT 7.470 (Katoh and Standley Citation2013) and phylogenetic analysis was conducted using the maximum-likelihood (ML) method with RAxML (Stamatakis Citation2014) pipeline available in the CIPRES Science Gateway (Miller et al. Citation2010). The ML tree was constructed using the general-time reversible (GTR) with gamma distribution (+G) (=GTR + G) nucleotide substitution model, with 1000 bootstrap replicates. Two species, Gossypium hirsutum (Malvaceae) and Eucalyptus grandis (Myrtaceae) were included as outgroups. The ML tree fully resolved G. walla in a clade with three species of Aquilaria (). The molecular placement of G. walla using the complete cp genome sequences appears to be in line with previously reported finding using the intergenic spacer region trnL-trnF (Eurlings and Gravendeel Citation2005) and the combined dataset of matK, rbcL, trnL intron, trnL-trnF, and psbC-trnS (Farah et al. Citation2018).
Figure 1. Maximum-likelihood tree constructed based on the complete chloroplast genome sequences of 12 plant taxa from the family Thymelaeaceae, with Gossypium hirsutum (Malvaceae) and Eucalyptus grandis (Myrtaceae) as outgroups. All branch nodes are indicated with bootstrap support values based on 1000 replicates.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at http://www.ncbi.nlm.nih.gov under the accession number MW557323. The associated BioProject, SRA, and BioSample numbers are PRJNA698718, SRX10001997, and SAMN17734768, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:39–40.
- Eurlings MCM, Gravendeel B. 2005. TrnL-trnF sequence data imply paraphyly of Aquilaria and Gyrinops (Thymelaeaceae) and provide new perspectives for agarwood identification. Plant Syst Evol. 254(1–2):1–12.
- Farah AH, Lee SY, Gao Z, Yao TL, Madon M, Mohamed R. 2018. Genome size, molecular phylogeny, and evolutionary history of the tribe Aquilarieae (Thymelaeaceae), the natural source of agarwood. Front Plant Sci. 9:712.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans (LA). p. 1–8.
- Pern YC, Lee SY, Ng WL, Mohamed R. 2020. Cross-amplification of microsatellite markers across agarwood-producing species of the Aquilarieae tribe (Thymelaeaceae). 3 Biotech. 3(10):1–9.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Subasinghe S. 2013. Agarwood resin production from Walla Patta (Gyrinops walla): the tree of the future. LINK Nat Prod Digest. 9:21–24.
- Subasinghe S, Hettiarachchi DS. 2013. Agarwood resin production and resin quality of Gyrinops walla Gaertn. Int J Agric Sci. 3:356–362.
- Subasinghe S, Hettiarachchi DS, Rathnamalala E. 2012. Agarwood-type resin from Gyrinops walla Gaertn: a new discovery. J Trop For Environ. 2:43–48.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- UNEP-WCMC (Comps). 2021. The checklist of CITES species website [Internet]. CITES Secretariat, Geneva, Switzerland. Cambridge: UNEP-WCMC; [cited 2021 Jan 26]. http://checklist.cites.org.
