Abstract
Brassica carinata A. Braun (Ethiopian rape), which was derived from the interspecific hybridization between B. nigra and B. oleracea, is used as both an oilseed and a leafy vegetable. The complete chloroplast (cp) genome of a purple B. carinata was obtained. This cp genome has a typical quadripartite structure and is 153,641 bp in length. The GC content of the cp genome is 36.39%. A total of 113 genes were predicted on this cp genome, including 79 protein coding, 4 rRNA, and 30 tRNA genes. Among these genes, 18 genes were duplicated (7 tRNAs, 4 rRNAs, and 7 protein coding genes). Sixty-eight SSR loci, including 11 compound SSRs, were identified in this cp genome by MISA. The phylogenetic tree analysis fully resolved B. carinata in a clade with B. nigra. This study provides important information for future evolution, genetic and molecular biology studies of B. carinata.
Brassica carinata, referred to by the common names Ethiopian rape, is an amphidiploid species derived from the diploid species B. nigra and B. oleracea (Nagaharu Citation1935). It is believed to have originated in Ethiopia (Gómez-Campo and Prakash Citation1999), where it is used as both an oilseed and a leafy vegetable. Seed oil from B. carinata has industrial applications due to its high erucic or linolenic acid contents (Warwick et al. Citation2006).
A deep purple B. carinata (accession BC-PL1) was gathered from Longwen mountain (26°23′11″N, 106°38′32″E), Guizhou province, China, and the voucher specimen was deposited in the herbarium of Guizhou Normal University (URL: https://sjxy.gznu.edu.cn/, Qun Feng, [email protected]) under the voucher number: ZB20201011. The total DNA of B. carinata was extracted by CTAB method, and the DNA libraries were prepared and sequenced using the Illumina HiSeq 4000 and PacBio Sequel platform (Illumina Inc. San Diego, CA). The cp genome was assembled using a de novo strategy according to the work of Du et al. (Citation2020). The annotated cp genome of B. carinata is publicly available in GenBank (accession number of MW628493).
The cp genome of B. carinata has a typical quadripartite organization and is 153,641 bp in length. The genome is composed of a large single-copy region (LSC, 83,561 bp), a small single-copy region (SSC, 17,696 bp), and two inverted repeats (IR, 26,192 bp). GC content of the genome is 36.39%. A total of 113 unique genes were predicted, including 79 were protein coding, 4 rRNA, and 30 tRNA genes. Among these genes, 18 genes were duplicated (7 tRNAs, 4 rRNAs, and 7 protein coding genes). Sixty-eight SSR loci, including 11 compound SSRs, were identified in this cp genome by MISA (Beier et al. Citation2017).
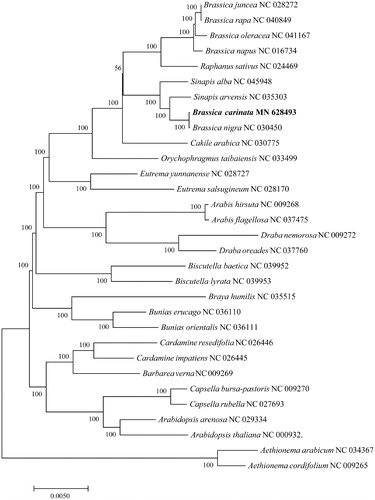
The phylogenetic tree was constructed based on complete cp sequences of B. carinata and other 30 related species in the Brassicaceae, and two Aethionema species were designated as the outgroup. These sequences were aligned using the default settings in MAFFT version 7 (Katoh and Standley Citation2013). The tree was constructed with the MEGA7.0 (Kumar et al. Citation2016) using maximum-likelihood (ML) optimality criterion with the Tamura-Nei nucleotide substitution model. The bootstrap analysis with 1000 replicates was used to confirm the stability of each tree node. B. carinata and B. nigra were fully resolved on a branch sister to other species of Brassica (). This result was consistent with the work of Li et al. (Citation2017), supporting the idea that the cytoplasm of B. carinata derived from B. nigra. The assembly of chloroplast genome sequence of B. carinata will be helpful for further study on its phylogenetic study, population genetics and breeding.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW628493. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA721808, SRR14235359, and SAMN18739861 respectively.
Additional information
Funding
References
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Du X, Zeng T, Feng Q, Hu L, Luo X, Weng Q, He J, Zhu B. 2020. The complete chloroplast genome sequence of Yellow Mustard (Sinapis alba L.) and its phylogenetic relationship to other Brassicaceae species. Gene. 731:144340.
- Gómez-Campo C, Prakash S. 1999. 2 Origin and domestication. In Developments in plant genetics and breeding. Elsevier; Vol. 4. p. 33–58.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li P, Zhang S, Li F, Zhang S, Zhang H, Wang X, Sun R, Bonnema G, Borm TJA. 2017. A phylogenetic analysis of chloroplast genomes elucidates the relationships of the six economically important Brassica species comprising the triangle of U. Front Plant Sci. 8:111.
- Nagaharu U. 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot. 7(7):389–452.
- Warwick SI, Gugel RK, McDonald T, Falk KC. 2006. Genetic variation of Ethiopian mustard (Brassica carinata A. Braun) germplasm in western Canada. Genet Resour Crop Evol. 53(2):297–312.