Abstract
Atractylodes japonica Koidz. ex Kitam. is a perennial herbal plant, and its dried rhizomes have been widely used as traditional medicine in China and Japan. In this study, we assembled and annotated the complete chloroplast (cp) genome sequence of A. japonica using the high-throughput sequencing approach. The cp genome of A. japonica is 153,208 bp in length with the overall GC content of 37.7%, including two inverted repeat (IR) regions of 25,147 bp, which was separated by a large single-copy (LSC) region of 84,255 bp and a small single-copy (SSC) region of 18,659 bp. 113 unique genes were annotated in the genome, including 80 protein-coding genes, 29 represented tRNA genes, and four denoted rRNA genes. A maximum-likelihood phylogenetic analysis with 38 complete cp sequences showed that Atractylodes formed a monophyletic clade, and A. japonica and A. koreana formed a subclade in Atractylodes. This study provides the chloroplast genome structure features and phylogenetic relationship of A. japonica.
Atractylodes japonica Koidz. ex Kitam. (Chinese name ‘Guancangzhu’) is a perennial herbal plant in the Atractylodes genus of Asteraceae family, which has potential medicinal value. The crude drug derived from the rhizomes of A. japonica has been used as the local medicinal ‘Cangzhu (So-jutsu, in Japanese)’ in Jilin Province of China (Rui and Chou Citation2020), however, it is not an official resource of ‘Cangzhu’ in Chinese pharmacopoeia for a long time (Commission Citation2020). In addition, the rhizomes of A. japonica have been classified into ‘Baizhu (Byaku-jutsu, in Japanese)’ in the Japanese pharmacopoeia (Kim et al. Citation2016). Although A. japonica has been treated as a synonym of A. chinensis according to the Flora of China (Linrong Citation1987), the mainly pharmacological active ingredients of the crude drugs are different. For instance, the content of atractylodin in A. chinensis is higher than A. japonica (Cho et al. Citation2016; Kim et al. Citation2018; Jeong et al. Citation2020). The purpose of this study was to analyze the structure of the complete cp genome of A. japonica and clarify its phylogenetic relationship with other species in Atractylodes.
Fresh leaves of Atractylodes japonica were collected from Tonghua City, Jilin Province (N41°43′, E125°56′). Its voucher specimen was deposited in the herbarium at the Chengde Medical University (http://www.cdmc.edu.cn/) with the voucher number as HPAB0017. Total genomic DNA was extracted from the leaf tissue using modified CTAB method (Porebski et al. Citation1997). The quantity and quality of the DNA were examined using Qubit 4.0 (Thermo Fisher Scientific Inc., USA). Purified DNA was used to construct sequencing library according to the TruSeq DNA PCR-free library preparation guide. The Illumina NovaSeq platform was employed to conduct high-throughput sequencing, and approximately 1.3 GB of raw data was generated with 150 bp paired-end read lengths. The sequencing adapter and low-quality reads were filtered using Trimmonmatic v0.38 (Bolger et al. Citation2014), and only the ‘paired’ output files were used for subsequent analysis. The complete chloroplast genome was assembled by the organelle assembly NOVOPlasty v4.2.1(Nicolas et al. Citation2017). The insert size was set to 350 bp and the complete cp genome of A. lancea (Accession number: NC_037483.1) was selected as a reference in the configure file of NOVOPlasty. The CPGAVAS2 (www.herbalgenomics.org/cpgavas2) (Shi et al. Citation2019) was used to annotate protein-coding, rRNA, and tRNA genes of cp genome and visualization with the default parameters. The final chloroplast complete genome of A. japonica was submitted to GenBank (Accession number: MW301112).
The chloroplast genome sequence of Atractylodes japonica was 153,208 bp in length and showed a conserved quadripartite structure like most angiosperm plants such as Withania somnifera (Mehmood et al. Citation2020) and Magnolia polytepala (Sun et al. Citation2020), with two reverse repeated regions (IRa and IRb) of 25,147 bp in length. The repeat regions divided the entire genome into two single-copy regions, namely a small single-copy region (SSC) and a large single-copy region (LSC) with 18,659 bp and 84,255 bp, respectively. The total GC content of the chloroplast genome was 37.7%. 113 unique genes were annotated in the genome, including 80 protein-coding genes, 29 represented tRNA genes, and four denoted rRNA genes (rrn23S, rrn16S, rrn5S, and rrn4.5S). Furthermore, 17 genes were annotated as containing introns, 10 (seven protein-coding and three tRNA genes) of which contained one intron and seven of which (rps12, ycf3, ndhB, rpl2, clpP, trnA-UGC and trnI-GAU) contained two introns. Moreover, petB, petD, and rpl16 have small exons, and the length of these small exons for the three genes are 6 bp, 8 bp and 9 bp, respectively. Finally rps12 was identified as a trans-splicing gene. In addition, there is another complete chloroplast genome sequence (Accession number: MT834523) of Atractylodes japonica can be found in Genbank submitted by Wang et al. (Citation2021), and there are lots of annotation errors in this released sequence. Four genes (psbL, ycf15, trnV-UAC and trnfM-CAU) failed to be annotated, three genes named atpB, ndhD and rpoC2 have been incorrectly annotated with wrong start or stop positions, 17 single nucleotide polymorphism (SNP) sites have been found in 13 protein-coding genes including atpA, atpF, ccsA, clpP, matK, ndhA, petB, psaB, rpl20, rpoB, rpoC1, rps19 and ycf2 comparing to the sequence reported in this study. These annotation errors and SNPs can be found in Supplementary Table 1.
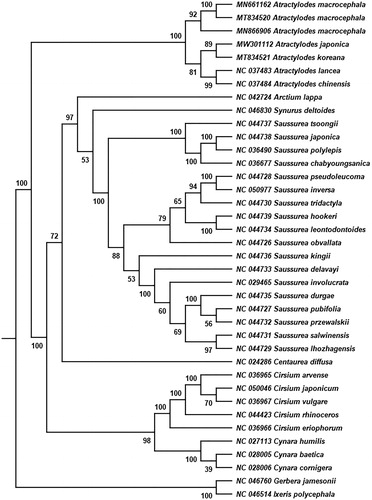
To confirm the phylogenetic location of A. japonica in the genus Atractylodes, a total of 38 complete chloroplast genomes belong to 9 genera of the Asteraceae family were used for phylogenetic analysis (Cai et al. Citation2020) based on the Maximum Likelihood (ML) method using RAxML v8.2.12 (Stamatakis Citation2014) with 1000 bootstrap replicates. Gerbera jamesonii and Ixeris polycephala were used as the outgroups for tree rooting (Susanna et al. Citation1995). The phylogenetic tree showed that Atractylodes formed a monophyletic clade with a bootstrap value of 100 (). A. macrocephala formed a branch independently with a bootstrap value of 92. A. chinensis and A. lancea formed a subclade in Atractylodes with a bootstrap value of 99, whereas A. japonica and A. koreana formed another subclade with a bootstrap value of 89. This study reports the chloroplast structure features of A. japonica, which provides valuable genetic information for its phylogenetic location in Atractylodes genus.
Disclosure statement
The authors reported no potential conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. MW301112. The associated BioProject, SRA, and BioSample numbers are PRJNA682118, SRR13181860, and SAMN16981139 respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Cai H, Chen C, Wang Y, Wang H. 2020. The complete plastome sequence of Atractylodes macrocephala (Asteraceae: Cardueae), an important medicinal plant in East Asia. Mitochondrial DNA Part B. 5(1):951–952.
- Cho H-D, Kim U, Suh JH, Eom HY, Kim J, Lee SG, Choi YS, Han SB. 2016. Classification of the medicinal plants of the genus Atractylodes using high-performance liquid chromatography with diode array and tandem mass spectrometry detection combined with multivariate statistical analysis. J Sep Science. 39(7):1286–1294.
- Commission CP. 2020. Pharmacopoeia of the People's Republic of China Part I. Beijing: China Medical Science Press; p. 168–169.
- Jeong J-T, Chung H, Ha B-K, Gil J, Lee J-H, Lee Y-J, Kim MR, Oh MW, Park CG, Chang JK, et al. 2020. Development of 18 microsatellite markers for Atractylodes japonica. Appl Plant Sci. 8(5):e11350.
- Kim J-H, Doh E-J, Lee G. 2016. Evaluation of medicinal categorization of Atractylodes japonica Koidz. by using internal transcribed spacer sequencing analysis and HPLC fingerprinting combined with statistical tools. Evid Based Complementary Altern Med. 2016:1–12.
- Kim J-H, Doh E-J, Lee G. 2018. Chemical differentiation of genetically identified Atractylodes japonica, A. macrocephala, and A. chinensis rhizomes using high-performance liquid chromatography with chemometric analysis. Evid Based Complementary Altern Med. 2018:1–16.
- Linrong S. 1987. Flora of China. Vol. 78. Beijing: Science Press; p. 23–29.
- Mehmood F, Abdullah , Shahzadi I, Ahmed I, Waheeda MT, Mirzaa B. 2020. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112:1522–1530.
- Nicolas D, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18–e18.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Rui M, Chou G. 2020. Three new polyacetylenes from Atractylodes japonica Koidz.ez Kitam [published online ahead of print, 2020 Nov 12]. Nat Prod Res. 1–8.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47:W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Sun L, Jiang Z, Wan X, Zou X, Yao X, Wang Y, Yin Z. 2020. The complete chloroplast genome of Magnolia polytepala: comparative analyses offer implication for genetics and phylogeny of Yulania. Gene. 736:144410.
- Susanna A, Jacas NG, Soltis DE, Soltis PS. 1995. Phylogenetic relationships in tribe Cardueae (Asteraceae) based on ITS sequences. Am J Bot. 82(8):1056–1068.
- Wang Y, Wang S, Liu Y, Yuan Q, Sun J, Guo L. 2021. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics. 22:103.