Abstract
Polytrichum commune, one of hair-cap mosses, is the type species of the genus Polytrichum Hedw. (Polytrichaceae). Here we present its complete plastome. The plastome of P. commune is successfully assembled from raw reads sequenced by HiSeq X ten system. Its total length is 126,323 bp consisting of four regions: large single copy (LSC) region (88,070 bp), small single copy (SSC) region (16,717 bp), and inverted repeats (IRs; 9,680 bp per each). It contains 128 genes (84 coding genes, eight rRNAs, and 36 tRNAs); nine genes (four rRNAs and five tRNAs) are duplicated in IR regions. The overall GC content is 28.9% and in the LSC, SSC and IR regions is 26.1%, 25.1%, and 45.5%, respectively. This plastome is an important sequence resource for further studies on the class Polytrichopsida.
Polytrichum commune Hedw., commonly known as a hair-cap moss, has been used as a traditional Chinese medicine as it has anticancer activity (Fu et al. Citation2009; Yuan et al. Citation2015). It shows clear differentiation of water-conducting tissue (hadrom and leptom), which is analogous to vascular tissue (xylem and phloem) of higher plants (Eschrich and Steiner Citation1968). Polytrichum is distinguished from the allied genus Pogonatum P. Beauv. by capsules with stomae (Smith and Gary Citation2007). Recent molecular phylogenetic studies presented that P. commune is a crown group of the family Polytrichaceae (Hyvönen et al. Citation2004), which is the sole member of the class Polytrichopsida. Till now, Pogonatum inflexum (Lindb.) Sande Lac. is the only available complete chloroplast genome of Polytrichopsida in GenBank. Polytrichum juniperinum Hedw. and Polytrichum strictum Menzies ex Brid. have only partial chloroplast genome data (de Freitas et al. Citation2018). Here, we present the plastome of P. commune as a first complete plastome of Polytrichum, the type genus of Polytrichaceae.
Polytrichum commune was collected in the tea farm of Fengyang Mountain, Zhejiang, China (27°52′46″N, 119°10′45″E). The specimen was deposited at the herbarium of East China Normal University (HSNU, http://museum.ecnu.edu.cn/; Rui-Liang Zhu, [email protected]) under the voucher number Zhu & Zhang 20200723-18. DNA was extracted using DNA Plantzol Reagent (Hangzhou Lifefeng Biotechnology Co., LTD). Genome sequencing was performed using HiSeq X ten system at BGI (Shenzhen), China, and de novo assembly was done by the GetOrganelle pipeline (Jin et al. Citation2020). The raw data were assembled using GetOranelle version 1.5.1 with the command get_organelles_reads.py. The command lines are as follows: get_organelle_reads.py −1 forward.fq −2 reverse.fq -o plastome_output -R 15 -k 21,45,65,85,105 -F plant_cp. The detailed steps to use getoranelle are shown on the website (https://github.com/Kinggerm/GetOrganelle). Geneious version 11.0.3 (Kearse et al. Citation2012) was used for plastome annotation, with Diphyscium foliosum (Hedw.) D. Mohr plastome (MN496311, Bell et al. Citation2020) as reference. CPGAVAS2 was used to further verify the tRNA genes (Shi et al. Citation2019).
The plastome of P. commune (GenBank accession MW528408) is 126,323 bp long (GC ratio is 28.9%) and has four subregions: 88,070 bp of large single copy (26.1%) and 16,717 bp of small single copy (25.1%) regions separated by 9,680 bp of inverted repeat (IR; 45.5%). It is longer than the sister species Pogonatum inflexum (MK131349, 125,415 bp) and contains 128 genes (84 protein-coding genes, eight rRNAs, and 36 tRNAs) and nine genes (four rRNAs and five tRNAs) duplicated in IR regions.
Sixteen complete chloroplast genomes () including P. commune were used for Bayesian Inference (BI, 2,000,000 generations, sampled every 1000 generations) and Maximum Likelihood (ML, bootstrap repeat is 1000) phylogenic trees using MRBAYES v3.2.7 (Ronquist and Huelsenbeck Citation2003) and IQ-TREE (Nguyen et al. Citation2015), respectively, after aligning whole plastome sequences using MAFFT v7.149b (Katoh and Standley Citation2013).
Figure 1. Maximum Likelihood (ML) and Bayesian Inference (BI) phylogenetic tree of 16 complete chloroplast genomes: Polytrichum commune (MW528408, in this study), Bartramia pomiformis (MT024676), Buxbaumia aphylla (MN496310), Diphyscium foliosum (MN496311), Fissidens nobilis (MK876184), Lewinskya incana (MK521877), Marchantia paleacea (NC_001319), Mnium marginatum (MT897999), Pogonatum inflexum (MK131349), Physcomitrella patens (NC_005087), Pseudocrossidium replicatum (MG132071), Sanionia uncinata (KM111545), Sphagnum palustre (KU726621), Takakia lepidozioides (AP014702), Tetraphis pellucida (KJ817846) and Tetraplodon fuegianus (KU095851). The ingroup consisted of 15 moss species representing 14 orders and five classes and Marchantia paleacea (NC_001319) as an outgroup. Phylogenetic tree was drawn based on ML tree. The numbers above branches indicate bootstrap values (BS) and Bayesian Posterior Probabilities (PP).
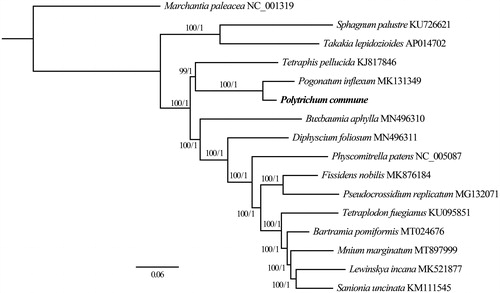
Phylogenetic trees show that class Polytrichopsida (P. commune and P. inflexum) is sister to class Tetraphidopsida (Tetraphis pellucida Hedw.), which is in accordance with previous studies (Volkmar and Knoop Citation2010; Liu et al. Citation2019) (). In addition, a basal clade was formed by Sphagnum palustre L. (Shaw et al. Citation2016) and Takakia lepidozioides S. Hatt. and Inoue (AP014702), which is same as the result of Cox et al. (Citation2004) and Qiu et al. (Citation2006), but is incongruent with Liu et al. (Citation2019). In summary, this suggests that additional bryophyte chloroplast genomes are needed to elucidate the phylogenetic relationships of these species. With the help of next generation sequencing technology, more and more plastome sequences of mosses will be published in the near future, which will allow us to have a better understanding of their phylogenetic relationships.
Disclosure statement
The authors are really grateful to the open raw genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data of Polytrichum commune that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW528408. The associated BioProject, Sequence Read Archive (SRA), and Biosample numbers are PRJNA698729, SRR13608611, and SAMN17734892, respectively.
Additional information
Funding
References
- Bell D, Lin Q, Gerelle WK, Joya S, Chang Y, Taylor ZN, Rothfels CJ, Larsson A, Villarreal JC, Li FW, et al. 2020. Organellomic data sets confirm a cryptic consensus on (unrooted) land‐plant relationships and provide new insights into bryophyte molecular evolution. Am J Bot. 107(1):91–115.
- Cox CJ, Goffinet B, Shaw AJ, Boles SB. 2004. Phylogenetic relationships among the mosses based on heterogeneous Bayesian analysis of multiple genes from multiple genomic compartments. Systematic Botany. 29(2):234–250.
- de Freitas KEJ, Metz GF, Cañon ERP, Roesch LFW, Pereira AB, Victoria FC. 2018. Characterization and phylogenetic analysis of chloroplast and mitochondria genomes from the Antarctic Polytrichaceae species Polytrichum juniperinum and Polytrichum strictum. Diversity. 10(3):89.
- Eschrich W, Steiner M. 1968. Die Struktur des Leitgewebesystems von Polytrichum commune. Planta. 82(1):33–49.
- Fu P, Lin S, Shan L, Lu M, Shen YH, Tang J, Liu RH, Zhang X, Zhu RL, Zhang WD. 2009. Constituents of the moss Polytrichum commune. J Nat Prod. 72(7):1335–1337.
- Hyvönen J, Koskinen S, Merrill GLS, Hedderson TA, Stenroos S. 2004. Phylogeny of the Polytrichales (Bryophyta) based on simultaneous analysis of molecular and morphological data. Mol Phylogenet Evol. 31(3):915–928.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu Y, Johnson MG, Cox CJ, Medina R, Devos N, Vanderpoorten A, Hedenäs L, Bell NE, Shevock JR, Aguero B, et al. 2019. Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat Commun. 10:1–11.
- Nguyen LT, Schmidt HA, von HA, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qiu YL, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al. 2006. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci. 103:15511–15516.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Shaw AJ, Devos N, Liu Y, Cox CJ, Goffinet B, Flatberg KI, Shaw B. 2016. Organellar phylogenomics of an emerging model system: Sphagnum (peatmoss). Ann Bot. 118(2):185–196.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Smith M, Gary L. 2007. Polytrichum commune. In: Flora of North America Editorial Committee, editors. Flora of North America. 27th ed. New York and Oxford: Oxford University Press.
- Volkmar U, Knoop V. 2010. Introducing intron locus cox1i624 for phylogenetic analyses in bryophytes: on the issue of Takakia as sister genus to all other extant mosses. J Mol Evol. 70:506–518.
- Yuan WJ, Cheng XX, Wang P, Jia YL, Liu QH, Tang W, Wang XB. 2015. Polytrichum commune L. ex Hedw ethyl acetate extract-triggered perturbations in intracellular Ca2+ homeostasis regulates mitochondrial-dependent apoptosis. J Ethnopharmacol. 172:410–420.
