Abstract
Strobilanthes biocullata is a plietesial species endemic to China. The complete chloroplast genome (cp genome) of S. biocullata was sequenced for the first time. The cp genome of S. biocullata is 144,012 bp in length. It consists of a large single copy (LSC) region (91,628 bp) and a small single copy (SSC) region (17,666 bp), which are separated by two inverted repeats (IRs, 34,718 bp). It contains 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The overall GC content is 38.2%. Phylogenetic analysis of 13 species has been conducted. This newly sequenced cp genome will be useful to further genetic diversity, phylogeny, and genomic studies of the genus Strobilanthes.
Strobilanthes Blume (Acanthaceae) is a genus containing about 450 species distributed in the tropical and subtropical regions of Asia with some species extending to Pacific Islands (Hu et al. Citation2011). Strobilanthes biocullata Y. F. Deng & J. R. I. Wood differs from all other species by its bracts having two swollen bulges on the dorsal surface and its plietesial life history which flowered gregariously after growing eight years and then died (Deng et al. Citation2010). In this paper, the complete chloroplast genome (cp genome) of S. biocullata was reported for the first time. The cumulative data have and will continue to contribute to our understanding of the cp genome feature of Strobilanthes.
The silica-gel dried leaves of S. biocullata were collected from Wudaoshui Zhen, Sangzhi Xian, Hunan Province, China (29°42′18.48″N, 109°55′28.03″E, 778 m). The voucher specimen (Yunfei Deng 27197) is deposited at the herbarium of South China Botanical Garden Herbarium, Chinese Academy of Sciences, Guangzhou, China. Total genomic DNA was extracted from leaves using modified CTAB method (Doyle and Doyle Citation1987). Short-insert (300–500 bp) libraries were constructed using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA) following the manufacturer's instructions. Pair-end (PE) sequencing was performed on the Illumina HiSeq 2500 instruments. To get plastid-like reads, the sequenced clean PE reads were filtered using GetOrganelle pipeline (Jin et al. Citation2018). The filtered reads were assembled using SPAdes v. 3.11.1 (Bankevich et al. Citation2012). The genome was automatically annotated using Plastid Genome Annotator (PGA) (Qu et al. Citation2019), coupled with manual correction in Geneious prime v.2020.0.5 (Kearse et al. Citation2012). Andrographis paniculata (GenBank accession number NC_022451) served as reference for assembly and annotation. The final complete cp genome was submitted to GenBank (accession number MW044601.1).
The complete cp genome of S. biocullata is 144,012 bp in length and presents a typical quadripartite structure including a large single copy (LSC) region (91,628 bp), a small single copy (SSC) region (17,666 bp), and two inverted repeat regions (IRs, 34,718 bp). The overall GC content of S. biocullata plastome is 38.2%. This plastome contains 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The ycf15 gene was considered as pseudogene due to the presence of internal stop codon.
To reconstruct the phylogeny of Acanthaceae, 13 Acanthaceae cp genomes were included and plastomes of two relative families were used as outgroups (). The sequences were aligned using MAFFT v. 7.450 (Katoh and Standley Citation2013). Maximum-likelihood (ML) tree was reconstructed using RAxML v. 8.2.11 (Stamatakis Citation2014) with the GTR + G+I nucleotide substitution model and all branch nodes were calculated under 1000 bootstrap replicates.
Figure 1. Maximum-likelihood (ML) phylogeny of Acanthaceae based on complete chloroplast genome sequences. Numbers at the right of nodes are bootstrap support values. GenBank accession numbers: Dicliptera acuminata MK830556.1, Rungia pectinate MK946456.1, Acanthus ebracteatus MT240944.1, Strobilanthes bantonensis MT576695.1, Sesamum indicum NC_016433.2, Andrographis paniculata NC_022451.2, Mentha spicata NC_037247.1, Strobilanthes cusia NC_037485.1, Echinacanthus attenuatus NC_039762.1, Aphelandra knappiae NC_041424.1, Clinacanthus nutans NC_042162.1, Justicia leptostachya NC_044668.1, Blepharis ciliaris NC_046601.1, Avicennia marina NC_047414.1, Barleria prionitis NC_048478.1, and Strobilanthes biocullata MW044601.1.
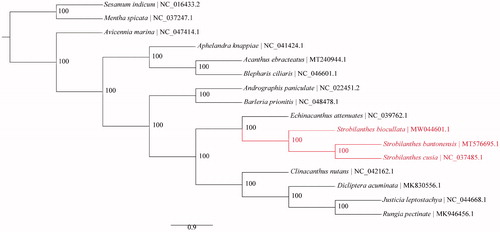
The results of the phylogenetics analysis confirmed the monophyly of Acanthaceae as previous studies (McDade et al. Citation2008) (). As expected, S. bantonensis and S. cusia were more close to S. biocullata. All the nodes received 100% bootstrap support. The genome data in this paper can be subsequently used for further genetic diversity, phylogeny, and genomic studies of the genus Strobilanthes and will contribute to further phylogeny studies of Acanthaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that newly obtained at this study are openly available in the NCBI (https://www.ncbi.nlm.nih.gov/) under accession number of MW044601.1.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Deng YF, Wood JRI, Fu Y. 2010. Strobilanthes biocullata (Acanthaceae), a new species from Hunan, China. Novon. 20:406–411.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hu JQ, Deng YF, Wood JRI. 2011. Strobilanthes. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China 19. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press. p. 381–429.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- McDade LA, Daniel TF, Kiel CA. 2008. Toward a comprehensive understanding of phylogenetic relationships among lineages of Acanthaceae sl (Lamiales). Am J Bot. 95(9):1136–1152.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
